Looking at the optic lobe tagging dashboard Codex it looks like many/several of the cells that is labeled in our farms of completed cells are missing in the dashboard for instance:
TM1 L have 655 in dashboard and 678 in farm, and
Tm9 L have 219 in dashboard and farm in 722
What could be the reason behind this? tag not registered in database or are the dashboard based on a older version of the database from codex?
I know that there were a few questions/comments brought up in another thread so I’m moving the responses/discussion over here.
Re: Differences in numbers between Flyer “Farms” in the Q&A Log and the Codex Tag Dashboard.
-
Most of the numbers are relatively close (+/- 20) to the numbers listed in the farms. This could be a difference in the parameters for the query run in Codex (it is based off of similarity in morphology, synaptic connectivity, input/output neuropils, etc.) to create the tagged/candidate lists. It could also mean that maybe some cells were prev. mislabeled or that the query missed some cells b/c they didn’t line up with the parameters. This is why we have the candidate lists, to help us find cells that have missing labels, may have gotten missed previously, and/or to correct any mislabels.
-
Codex is versioned data so it is possible that some cells have been updated since the “% tagged” lists were generated. The retinula cells (R1-6 & R7/8) are a prime example of a list that needs to be updated since the last Codex version at the end of April – there should be a great many more in the tagged list!
Re: Candidate list updating
- While the lightbulb status in FlyWire updates within a few minutes of submitting a label/tag, please be aware that Codex only updates approx. twice daily with the tags.
Re: Preventing Overlap of Tagging
-
The Tag Dashboard is a beta version and we are developing methods to help prevent users from checking the same batch of listed cells (Update here!)
-
Checking the lightbulb status is the most efficient way to confirm labels; but please be aware that some candidates may be in the list b/c they might be mislabeled! You can filter for unlabeled candidates on the list by adding
{and} {not} labelto end of the text in the search bar and hitting search. -
I recommend tagging the (now) left side of the Optic Lobes. Unsurprisingly, it is the side with the lowest percentage of tags. It is also the area with the least overlap with other users.
Re: Dm8s
- A Princeton lab tracer is currently focusing on the Dm8s
As for the tagging in the left lobe. I want to do at least some more Tm and TmY in the right lobe and only then move to the left one. I hope, that’s ok.
Perfectly fine! I’m not aware of anyone in the lab tagging any Tm or TmY cells, so they’re fair game ![]()
An FYI that I am gardening and tagging the (now left) Lobula Plate. Primarily focused on the TM4/TM5 cells right now but tagging others as I come across them. I haven’t updated my farms in the Google Sheet yet but so everyone knows where I’m actively working.
I’m working my way through the L1-5’s in the (new) left lobe atm, starting with 1’s. Keeping farm links up to date as i go. Will go back to the right lobe after to catch any that were missed.
Thanks all for posting and joining in on the tag party! ![]()
@st0ck53y No need to worry too much about the L1-5s on the (now) right OL. In the lab, twister (yes, originally from Eyewire!) is going through to catch any missed ones on that side.
Update: Candidate lists on Codex Tag Dashboard will now randomly shuffle
To prevent everyone from checking the same first 10 cells in a candidate list, the list order of the cells will shuffle every time you open a new instance of the candidate list for a cell type. This is great if you’d like to just pop in and tag a few cells for a short period of time ![]()
If you’d like to keep a stable candidate list, you can download the Cell IDs text list.
After a cell is labeled, it will be removed from the candidate cell list when Codex is updated (~ once daily).
All L1s in left lobe are id’d ![]() (except 2 maybes and one not yet traced through a black wedge). 789-791 cells, the ones along the edges are definitely the most difficult, and lots of missing somas ;~;
(except 2 maybes and one not yet traced through a black wedge). 789-791 cells, the ones along the edges are definitely the most difficult, and lots of missing somas ;~;
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6345892573478912
This is awesome @st0ck53y! ![]()
For the area with the detached lamina, did you tag the terminal arborization in the medulla for those that were missing the upper lamina/soma connections?
Yeah, one or two managed to cross the gap, but the rest all got tagged as far up as it could go ![]()
Just fyi, Codex is now being updated twice daily with new tags!
Hi Flyers,
Thank you so much for contributing to labeling the Optic Lobe cells ![]() ! I have a few updates regarding the current “rally” towards labeling cells. Here’s some of the latest awesome news:
! I have a few updates regarding the current “rally” towards labeling cells. Here’s some of the latest awesome news:
- we are currently focusing on labeling all the cells in the right Optic Lobe
- as of posting today, 34,515 (91%) neurons in the right optic lobe have been typed in the OL Catalog
The Tagging Dashboard has been retired. There have been some rapid algorithmic developments made for curating predicted/candidate cell type lists and we are now using the Optic Lobe Catalog.
You can also find the Optic Lobe Catalog and some additional tools for finding similar cells to label by clicking on the Research Beaker icon in Codex next to the “More” tab.
The “Optic Lobe Catalog” link is at the top of the dropdown menu.
If you are interested in using the Catalog, you’ll find that there are two options of finding lists of cells that could be labeled:
-
Lists of “Unknown” cells that are predicted to be certain cell types
-
Individual prediction lists by cell type (these are the Green
 colored links in the bulleted list under a cell type - note not every cell type will have a predicted list)
colored links in the bulleted list under a cell type - note not every cell type will have a predicted list)
If you’re really passionate or knowledgeable about a particular cell type, you may want to choose Option 2!
For Option 1, to help verify that a predicted cell is correct, there is now a link provided to the any previously labeled “source” cells of that cell type:
-
Select any individual cell from the predicted list to open up it’s Cell Info page.
-
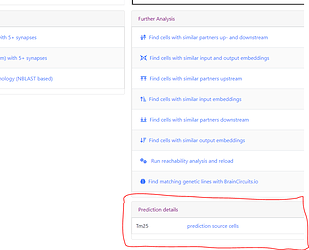
On the right side of the page, scroll down past the “Further Analysis” section. You’ll see a section titled “Prediction Details”:
-
Click on the Prediction Source Cells link.
-
This will open up a list with your predicted cell at the top and followed by any of the “source” cells that led to this cell being predicted to be a certain cell type.
-
Copy the IDs and compare away!
A few notes about the catalog:
-
Tm and TmY type cells are a great place to focus if you’re looking for a place to start and feel comfortable labeling them!
-
Pink links for individual cell types are lists of that cell type that are NOT in the Right Optic Lobe
-
You may see some new cell names with a different format:
- Any cell that has a number over 100 is a “provisional new cell type” ie. Mi104_1468
- The numbers after the _ are the neuropil layers where the cell has arbors (or innervated)
-
- For example, Mi104_1468 has arbors in medulla layers 1,4, 6, and 8
-
If you find a group of cells that don’t match any the cell types listed in the catalog and think that it needs a new provisional label, please email us a link to the group!
Happy tagging! ![]()
![]()
![]()
-M.
Forgot to share that in addition to the Reference for Layers in the Medulla, we now have the approx. Lobula layers marked out in this link here (includes the Medulla layers too): Lobula and Medulla Layers
This is helpful for determining cell type and comparing to the figures from Fischbach and other papers. Of course, if you need the approx. Lobula Plate layers, here’s KK’s handy reference using the Horizontal and Vertical System cells: Lobula Plate layers
Hi All,
Quick update the Optic Lobe Catalog is officially live for all folks on Codex. You can now find the Catalog for all the intrinsic cells in the right Optic Lobe under the “More” section on Codex.
As I mentioned on Twitch Office Hours today, we are still working through analysis and tagging straggler cells in the right Optic Lobe in the lab. If any Flyers are interested in helping us review, complete and tag some of these outlying cells, we have a few mini-missions for you!
Most of these cells are uncompleted due to missing somas so if you are comfortable and familiar with identifying any of the cell types, feel free to respond to this post and I can give you a list of predicted candidate types of that cell.
Cell Types include: Mi4, Mi8, Tm1, Tm2, Tm4, Tm9, Tm20, Dm8a/b, T1, T5a-d, C2, C3
For these cells, it is OK to complete the cell even if the soma is missing/cannot be found and the cell has the majority of its identifying characteristics (see Optic Lobe Catalog for similar sample cells to compare to) .
Thank for all of your contributions to labeling the optic lobe cells!
Cheers,
M.
I can work on some of these. Currently I’m working on Tm32 in the left OL and was planning to further split the Dm3 there, but these 2 can wait.