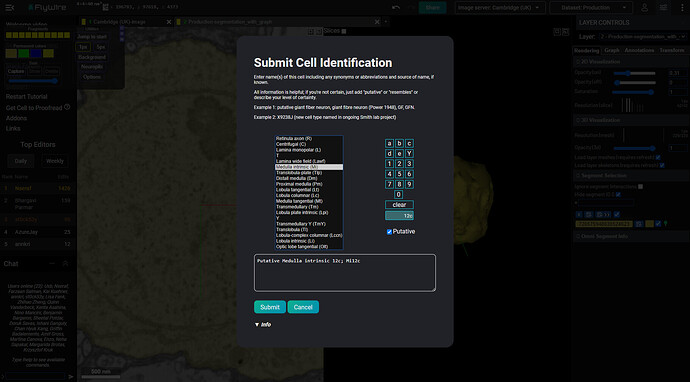
I’m currently working on an addon called “Cell Identification Helper” which should help with inputting correct names of cells during the Cell Identification process. The work is in progress, so there isn’t currently any link available for installing it. However, I’m starting this topic to ask you for your opinions and your help.
Here’s how it currently looks:
It working is simply - you select a “family” of a cell, then select it’s number and a suffix, if needed. You can also check the “Putative” checkbox to add the word before the whole name.
Here are some questions for you:
- What do you think about the UI/UX of it (how does it look, how to use it)?
- The current list on the left is copied from @Nseraf’s list in the Q&A gsheet:
a. Is the list complete?
b. Should it be in alphabetical or any other order, than it currently is?
c. Should it display both long and short names or only short (maybe only long)? - Should I add other exisitng long and/or short names for already existing cells?
a. If so, should the alternative names also be displayed in the list or only in the target text? - Should I add an author and a year for all the cells?
a. Does it should look like (Fischach et al., 1988) or maybe other format, maybe add other authors too?
b. Should I add initial(s) for autors’ name(s)? - Should I also attach a DOI identificator for all the known cells? (after all, one author can have multiple papers in a single year)
a. If so, what would be the preferent format? - How does the final name should look like (currently it is like on the picture)?
- If there are more than one long and short names, should I display them like that:
- long_name1; long_name2; short_name1; short_name2
or like that: - long_name1; short_name1; long_name2; short_name2
- a different way
As for a list of authors and DOIs, I’ve just started making such list. So I’ll try to do as many as possible and then ask for your help for the cells, I won’t be able to find sources.