This is great and definitely does bring up a good issue about naming and cell ID. Personally I think it may be best if we add a citation for the morphology if it’s not Fischbach. Fischbach seems like the generally agreed source/standard that everyone has been working from, but obviously there have been some different approaches since.
Thanks.
I’m currently working on an addon that will allow to add cells’ names easier during identification process. I’ll soon open a new thread for discussion, for how the names should look like.
An update to this while I was researching cell name types: Loi is just another term for Lobula intrinsic (instead of Li). See mentions in ex: https://www.cell.com/current-biology/pdfExtended/S0960-9822(17)30012-X
Medulla stratum, R7, R8, L1, L2
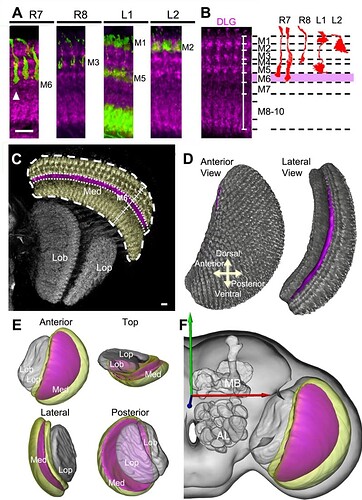
Generation of medulla volume models. A: Arborizations of retinotopic afferents (green) within medulla columns in relation to DLG-immunostained medulla strata (magenta). A long-form R7 (arrowhead) defines the proximal boundary of M6. B: Identification of specific strata in the distal medulla (M1–M6) based on differences in DLG-immunostaining intensity (left), using specific columnar neurons as references (right). C: Segmentation of the M6 stratum (magenta) and medulla (yellow) from the DLG-immunostained optic lobe (gray). D: Volume rendering of the M6 stratum (magenta) within the medulla (gray). E: Spatial relationship of M6 (magenta), medulla (yellow), lobula, and lobula plate (gray) volume models. F: The M6 volume model provided by the standard model fly brain. Med, medulla; Lob, lobula; Lop, lobula plate; MB, mushroom body; AL, antennal lobe. Scale bars = 10 μm.
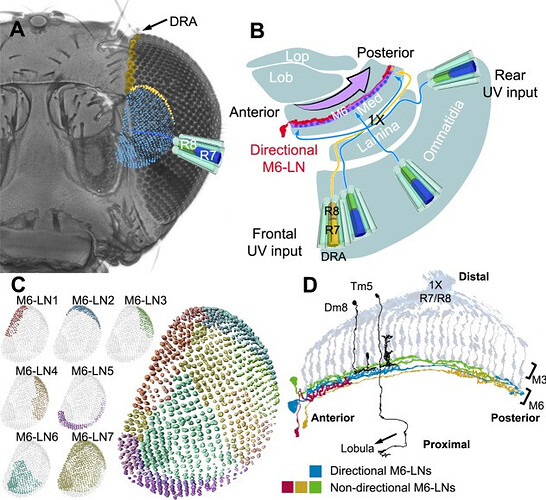
Schematic representation of UV vision in the M6 stratum. A: Retinotopic representation of UV-sensitive neurons in the M6 stratum. Each of approximately 800 R7 neurons relays UV information along one column to terminate at the M6 stratum, forming a UV retinotopic map (blue). The UV polarization-detecting DRA neurons (yellow) project to the dorsal-lateral M6 stratum. B: Top view of the left optic lobe shows chiasmatic information relay (×1) from frontal and rear ommatidia to posterior and anterior M6, respectively. Directional M6-LNs (red) relay information from the anterior to the posterior M6 (arrow). Med, medulla; Lob, lobula; Lop, lobula plate. C: Innervation territories of nondirectional M6-LN clusters (shown in different colors) form a mosaic of seven domains, covering the whole M6 retinotopic field. D: Functional motif in the M6 stratum: R7/R8 input neurons, narrow-field output projection neurons (black; i.e., Dm8 to the M4 stratum and Tm5 to the lobula), nondirectional LNs (green, red, and orange), and directional M6-LNs (blue).
Both images from: Diversity and wiring variability of visual local neurons in the Drosophila medulla M6 stratum - PMC
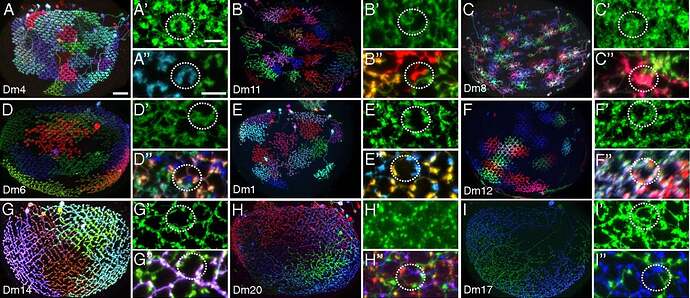
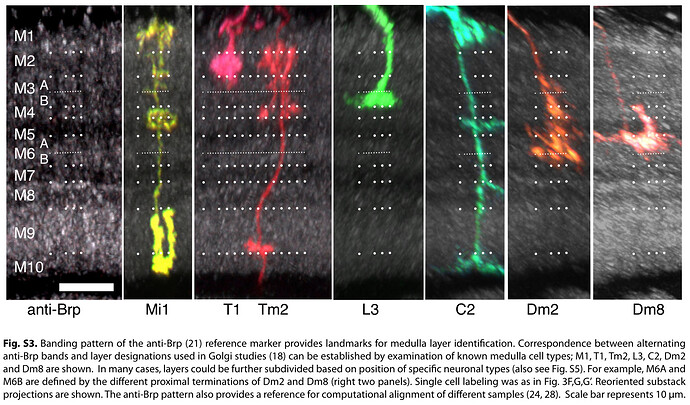
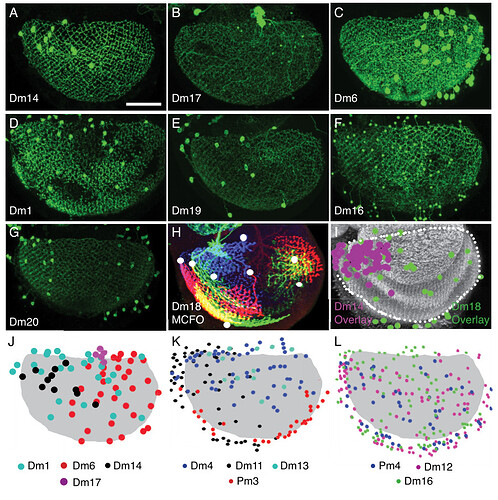
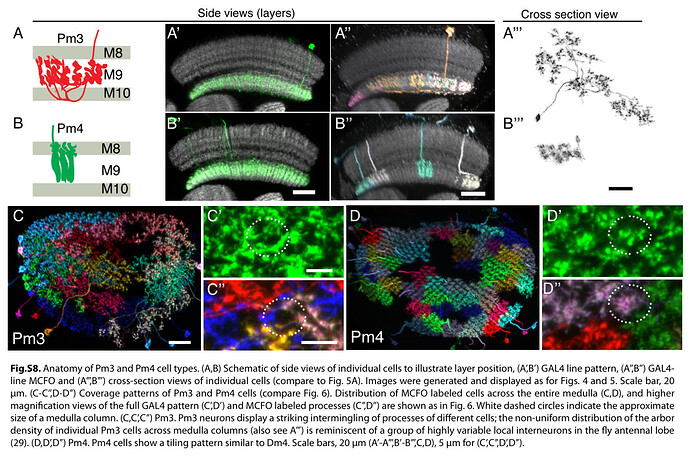
The first image helps with identifying layers in the Medulla. As one can see (C), there’s a recess around the Medulla (also visible in our rendered neuropils, no to much in the 2D view, but you can still see the layers). Above that is the M6 layer, as marked on the picture. The layer below is M7. It’s sometimes called serpentine layer. It separates Outer (Distal) Medulla from Inner (Proximal) Medulla. So, Dm cells should be above the recess, and Pm below it (closer to Lobula).
FlyLight | Janelia Research Campus a researcher just asked how to find neurons by name from this site in fw. lol tyty sharing this here if it’s not already. hehe
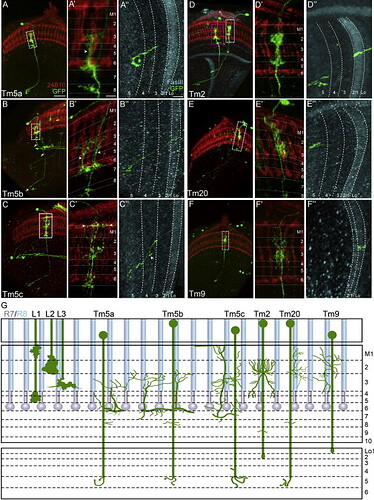
Different subtypes of Tm5:
Source: The Neural Substrate of Spectral Preference in Drosophila: Neuron
That’s probably the reason, why there are 4 slightly different Tm5’s in the Dischbach’s paper.
Keywords: Tm5a, Tm5b, Tm5c, Tm2, Tm9, Tm20
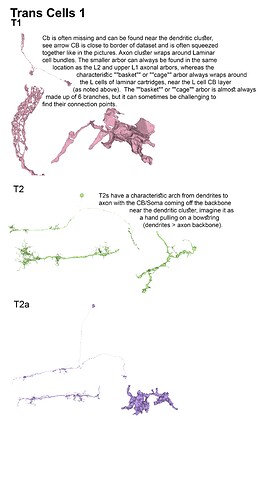
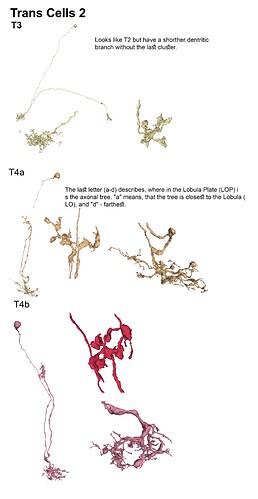
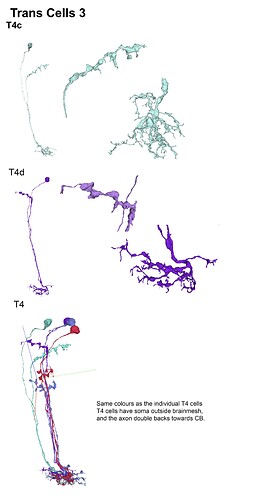
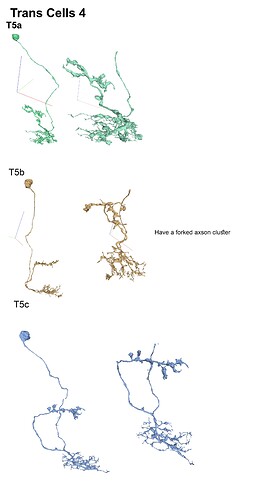
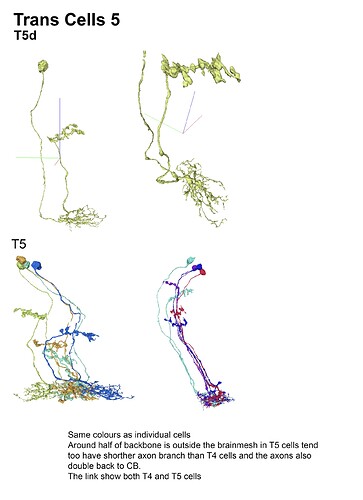
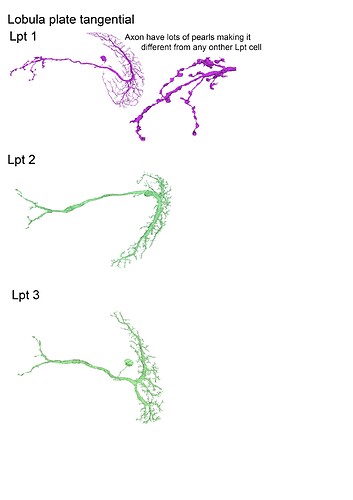
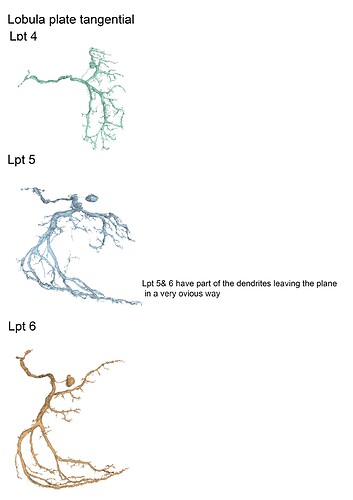
With the trans cells in gsheet finished here’s the illustrations: (I broke them up in 5 files so that 1 file wouldnt become non-printable by matter of resolution (ie: 200 x 40000 lol (bigger than A4))
Feel free to print etc away ![]()
These are finished as well, although I am not sure if we’ll get to identify any of these as I think they’ve already all been completed and identified by the lab ppl, but let’s have the visuals lol.
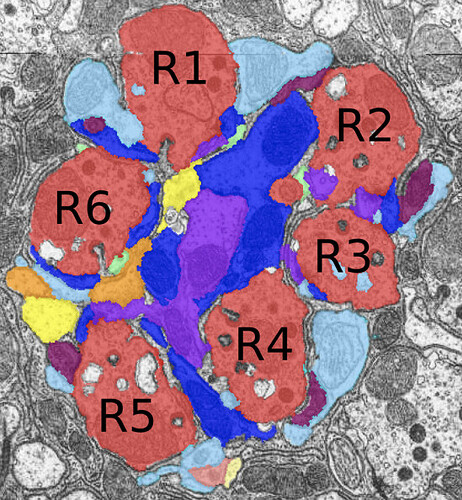
Q: Any way to tell R1 from other R# (other than 7/8s) in fw?
A: (From Emil Kind in youtube vid of yday’s symposium): “Very good question! It should be possible, but it is not trivial. The outer photoreceptors are arranged in each lamina cartridge according to a stereotyped pattern. This pattern is mirrored along the equator for the dorsal and ventral halves of the lamina. By identifying all outer PRs from a series of adjacent cartridges, one should be able to infer their R1-R6 fate.”
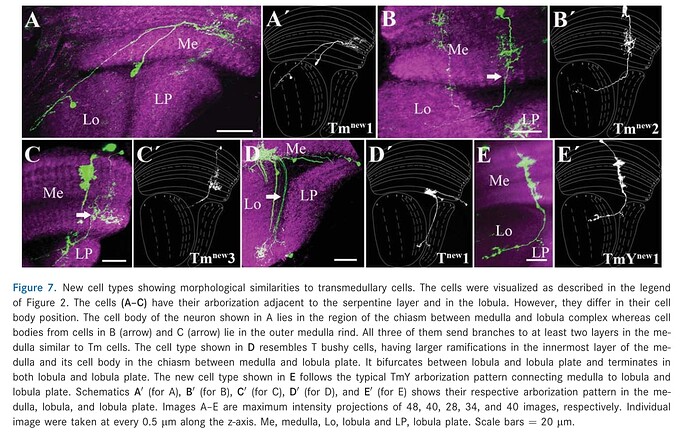
Keywords: Tmnew1, Tmnew2, Tmnew3, Tnew1, TmYnew1
Tmnew1 = ChaTmnew1,
Tmnew2 = ChaTmnew2,
Tmnew3 = ChaTmnew3,
Tnew1 = ChaTnew1,
TmYnew1 = ChaTmYnew1.
The other names are taken from FlyBase
Source: Neurons with cholinergic phenotype in the visual system of Drosophila - PubMed
Figure 1. A Lamina Cartridge in Drosophila melanogaster Reconstructed in Three Dimensions from Serial Electron Microscopy Images
(A) Single electron microscopy (EM) image in which the cell profiles have been segmented and digitally labeled with different colors.
(B) Three-dimensional shapes of the neurons in the reconstructed cartridge; see Figure S1 for shapes of individual neurons.
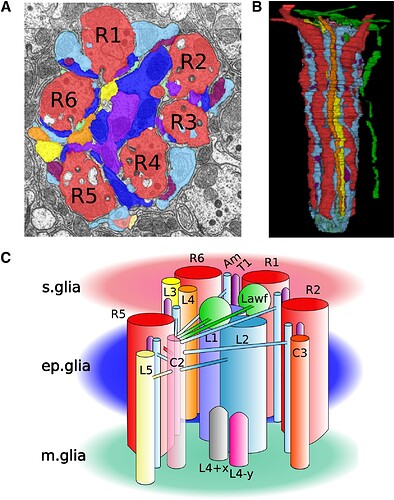
(C) Schematic version of (B). Neurons connect with lateral branches, here illustrated for neuron C2. The following abbreviations are used: R1–R6, photoreceptors terminals 1 to 6; L1–L5, lamina monopolar cells 1 to 5; L4+x and L4−y, incoming L4 collaterals from the two neighboring anterior cartridges along the +x and −y axes, respectively; Am, amacrine cells; T1, T medulla neuron 1; C2 and C3, centrifugal medulla neurons C2 and C3; Lawf, lamina wide-field cells; ep. Glia, epithelial glia; s. glia, satellite glia; m. glia, marginal glia. To enable their visualization, we show only four of the six photoreceptors, five of the six T1 branches, five of the six amacrine vertical branches, and two of the 11 Lawf branches.
Figure 2. Connectivity in the Lamina Cartridge
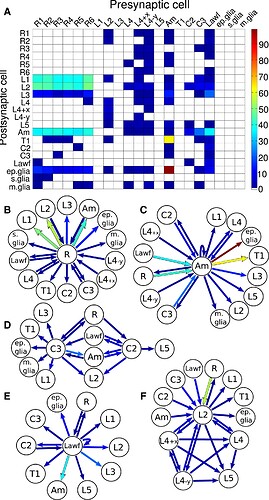
(A) Connectivity matrix; see Table S1 for an extended matrix including neuronal subtypes and Table S2 for a numerical version. Hotter colors indicate a larger number of synapses.
(B–F) Representation of neurons and connecting neighbors for a photoreceptor (B), an amacrine neuron (C), centrifugal neurons C2 and C3 (D), a large wide-field neuron Lawf (E), and large monopolar cell L2 (F).
From Marta Rivera-Alba, Shiv N. Vitaladevuni, Yuriy Mishchenko, Zhiyuan Lu, Shin-ya Takemura, Lou Scheffer, Ian A. Meinertzhagen, Dmitri B. Chklovskii, Gonzalo G. de Polavieja,
Wiring Economy and Volume Exclusion Determine Neuronal Placement in the Drosophila Brain,
Current Biology,
Volume 21, Issue 23,
2011,
Pages 2000-2005,
ISSN 0960-9822,
https://doi.org/10.1016/j.cub.2011.10.022.
(Wiring Economy and Volume Exclusion Determine Neuronal Placement in the Drosophila Brain - ScienceDirect)
T4, T5 and their synaptic partner neurons.
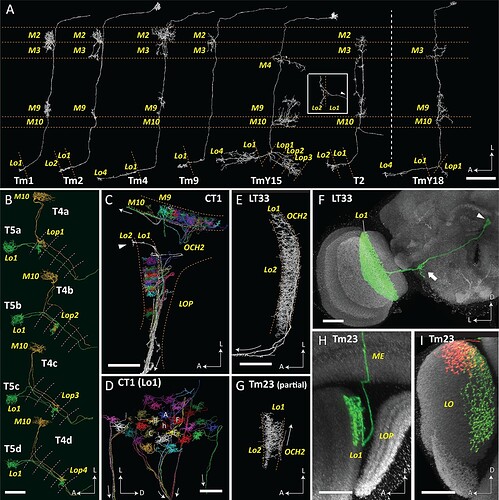
(A) T5 forms synaptic contacts with Tm1, 2, 4, 9, TmY15 and T2 cells. T4 is presynaptic to a newly identified TmY18 cell in M10. These synaptic partners of T5 and T4 have cell bodies in the medulla cortex distal to stratum M1, except T2 has a soma between the medulla and lobula. Tm and T2 cells receive inputs mainly in the medulla and project to the lobula, whereas TmY cells project to both the lobula and the lobula plate. All reconstructed cell profiles closely capture the profiles of those reported from Golgi impregnation (Fischbach and Dittrich, 1989) except the lobula terminal of T2 (inset), shown here from a different angle (lateral view, arrowhead indicates the outgoing fiber connected to the medulla and cell body). (B) Paired T4 and T5 cells. T4/5a, b, c, and d cell pairs project to the lobula plate, to strata Lop1, 2, 3 and 4, respectively. Their cell bodies are located in the lobula plate cortex, posterior to the lobula plate. (C, D) Parts of CT1 reconstructed in both the medulla and lobula reveal the morphology of its color-coded columnar terminals. Fibers connecting the terminals enter the second optic chiasm (OCH2) before exiting the optic neuropils (trajectories shown by arrows). Most branches terminate in medulla stratum M10 or lobula stratum Lo1, but a few terminals project deeper, into Lo2 (arrowhead in (C)). Columnar terminals in Lo1 corresponding to a home (h) column and the surrounding six columns (A–F) are indicated in (D). (E) A partial reconstruction of LT33, a novel neuron in lobula stratum Lo1. (F) Optic lobe and central brain projection pattern of an LT33 cell visualized using multicolor stochastic labeling (MCFO; see Materials and methods). The arrowhead indicates the cell body, and the arrow shows the site of branching in the posterior lateral protocerebrum. (G) Partial reconstruction of Tm23 in stratum Lo1. (H, I) Tm23 cells and their projection patterns in the optic lobe visualized using MCFO. Images show two different views generated from the same confocal stack. Images in (F), (H), and (I) show resampled views generated from confocal stacks using Vaa3D. For clarity, the images in (F) and (H) were segmented to omit additional labeled cells (distinguished by labeling color and position) that are also present in the displayed volume. In (I), red and green signals each represent single Tm23 cells. Scale bars: (A) 20 μm; (B), (C), (E), (G) 10 μm; (D) 5 μm; (F), (H), (I) 20 μm.
Citation:
Kazunori Shinomiya, Gary Huang, Zhiyuan Lu, Toufiq Parag, C Shan Xu, Roxanne Aniceto, Namra Ansari, Natasha Cheatham, Shirley Lauchie, Erika Neace, Omotara Ogundeyi, Christopher Ordish, David Peel, Aya Shinomiya, Claire Smith, Satoko Takemura, Iris Talebi, Patricia K Rivlin, Aljoscha Nern, Louis K Scheffer, Stephen M Plaza, Ian A Meinertzhagen (2019) Comparisons between the ON- and OFF-edge motion pathways in the Drosophila brain eLife 8:e40025
I have problems with identifying all the Mt cells. Especially, that they are spreaded all over the Fischbach’s paper. So I’ve decided to put them all in one place and remove all the other cells and layers from the picture. Here is the result:
There’s missing Mt6, because it isn’t in the paper as a sketch. It is in the further part of the document, but only as a not very clear microscophic photo. There also photos of Mt13, Mt14 and Mt15, but all harder to guess their shape.
FlyBase informs about 4 other Mts, all identified by their neurotransmitters.
Thanks! I added this to the cell deck
Some more images from the same source as in the first post of this thread, and its appendix:
(the dots mark positions of the somas)
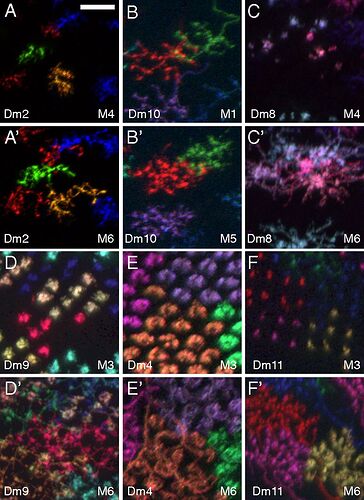
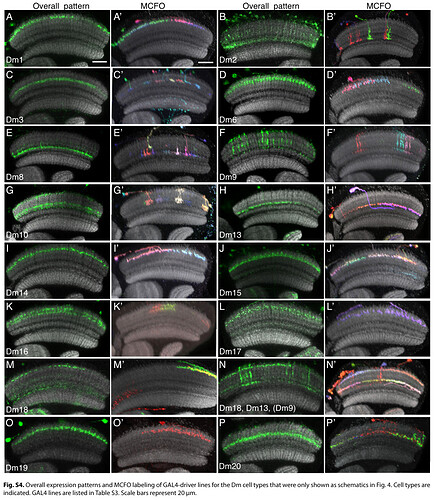
Keywords: Mi1, T1, Tm2, L3, C2, Dm1, Dm2, Dm3, Dm4, Dm6, Dm8, Dm9, Dm10, Dm11, Dm12, Dm13, Dm14, Dm15, Dm16, Dm17, Dm18, Dm19, Dm20, Pm3, Pm4
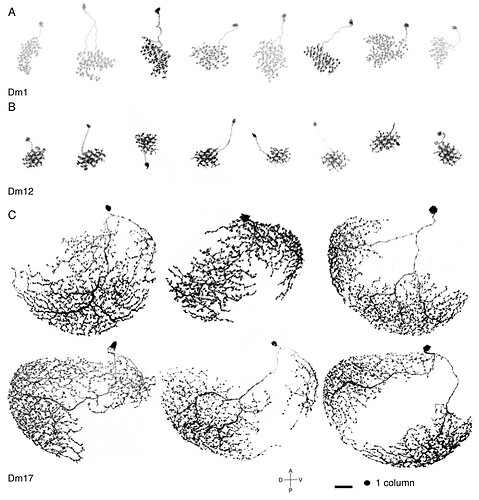
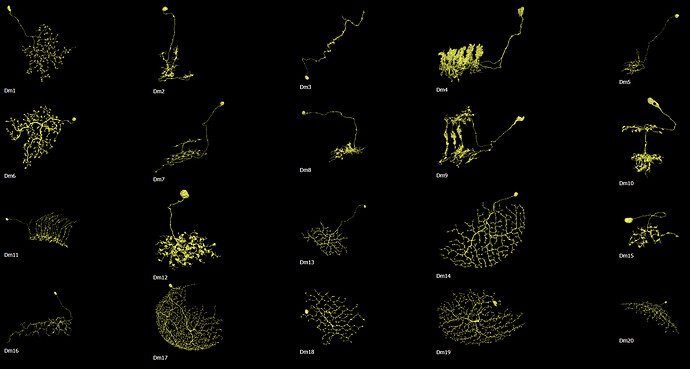
I had trouble recognizing, which Dm is which, so I made this collection of the basic 20 types:
Cells are NOT up to scale to each other, because the differences would be to big and some smaller cells wouldn’t be recognizable.
Great visual, I added to the cell guide
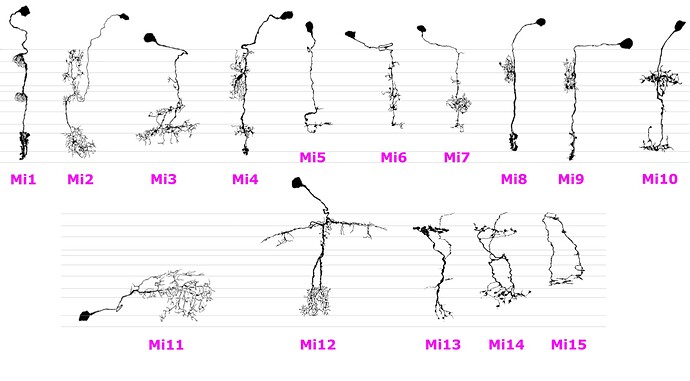
I’ve collected all the Mis from the Fischbach’s paper, scaled them, rotated, removed the original layers and put side by side in their respective layers.
Also added Mi13, Mi14 and Mi15 from this paper. These have inverted colors and set thresholds to make them black&white.
Here’s the result: