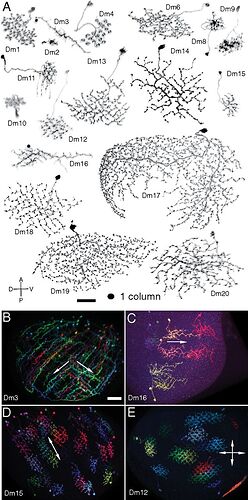
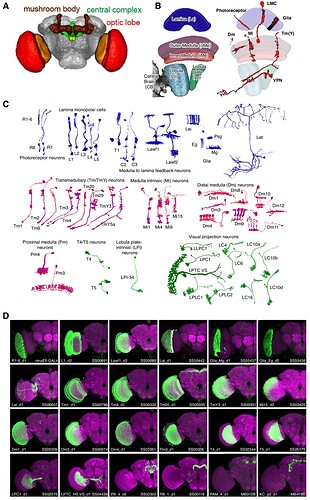
I use the conventions HQ used in the Optic Cell slideshow - Optic Lobe Cell Name Guide - Google Slides - which I think all follow Fischbach.
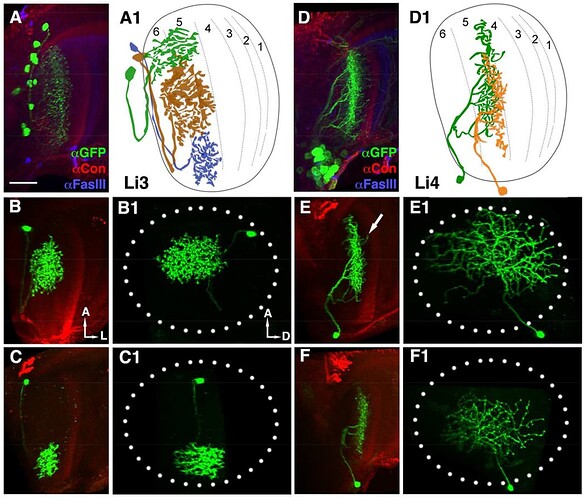
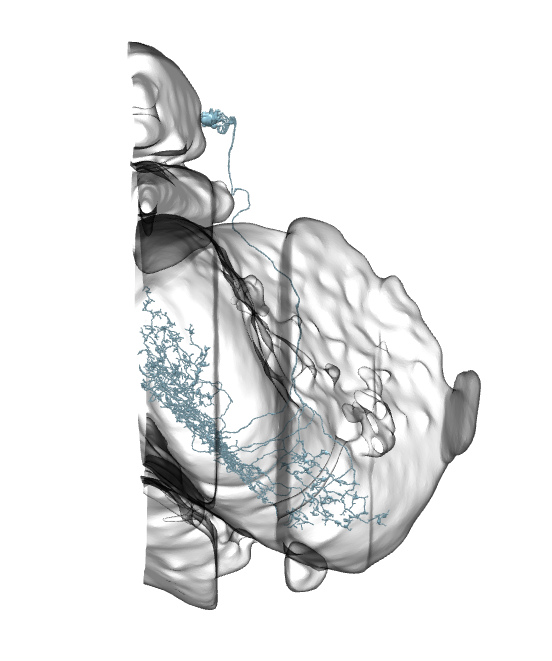
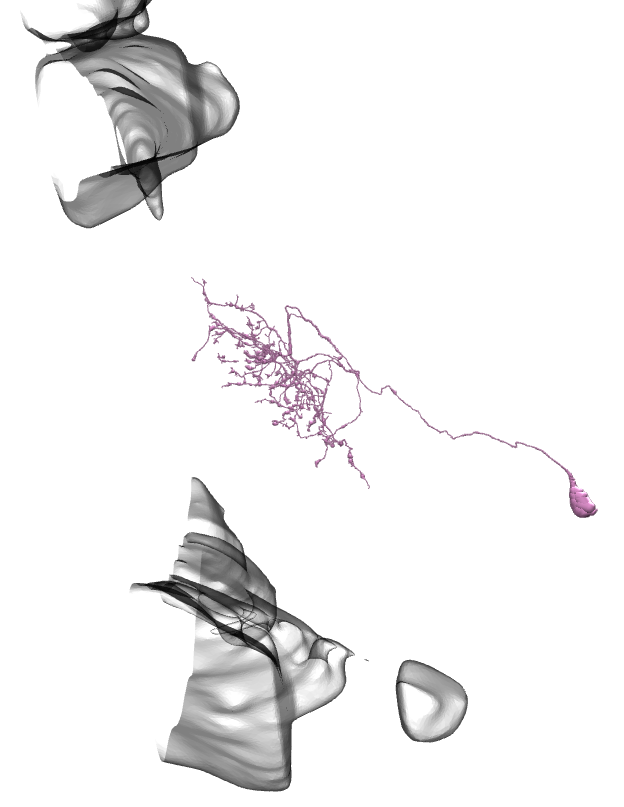
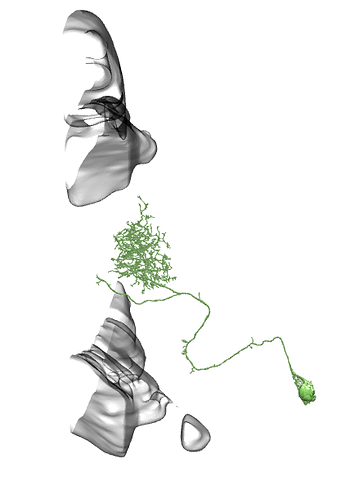
Li3 and Li4 shown. Source.
In each optic lobe, about 12 Li3 neurons elaborate dendritic arbors spanning the entire lobula strata 5 and 6 (A). Approximately 30 Li4 neurons, clustered in the posterior lobula cortex have dendritic arbors covering the entire lobula stratum 5.
Made a few examples on placement in the optic lobe for C and L cells with different settings see excel sheet
- different oppasity 0,1- 0,05- 0,02
- the whole lobe or just relevant parts
- all neuropils or just medulla
- naming the neuron black/red/none
- neurons in colour or black (more work)
- standard orientation (most pictures) or different orientations?
Before doing anything more on this i would like to know if this is useful or not.
thats nice! idk if it needs more options (like neurons in black etc), what could be useful maybe would be to pair diff types that synapse, as in “if you’re searching for (example) L2 it synapses with LAWF (idk that it does just exampling 2 diff types), so if you can’t find LAWF CB/ L2 CB layer if you find the other just search synapsing boutons for a LAWF/L2”?
The different options was just to get some respons on what you think looks best, would only be one picture at the end.
who the different neurons synapse with/ are togheter with would be a lot of work and not something i am prepered to take on at this time.
I see, sry misunderstood, then diff parts and whole brain, all neuropils and neuron names id see as something useful, maybe diff orientations but not sure on that one.
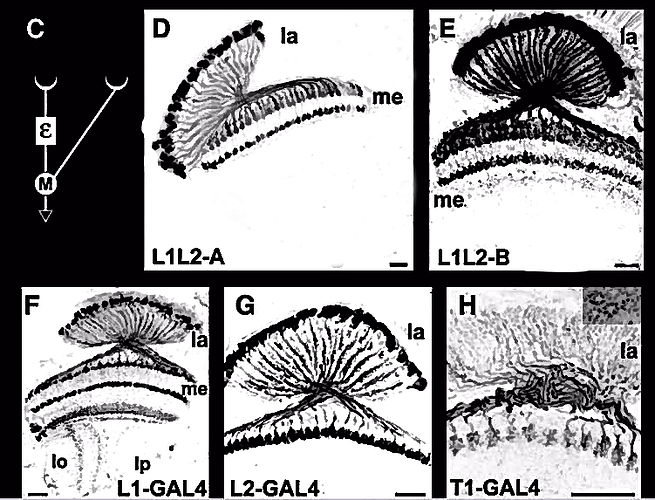
Attempted an inversion and clean up of the image from this 2007 paper in hopes of improving visual contrast and acuity.
L1L2-A, L1L2-B, L1-GAL4, L2-GAL4, T1-GAL4
Attempting a new type of illustration with an emphasis on the silhouette ala Fischbach, highlighting cluster shapes and optic lobe locations. Should I continue with these?
Edit: v2 with edits suggested by annkri -
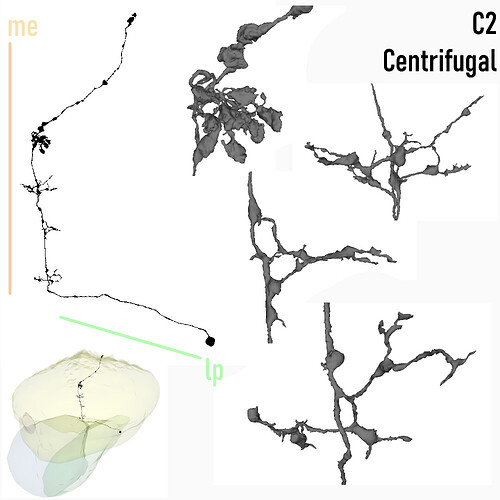
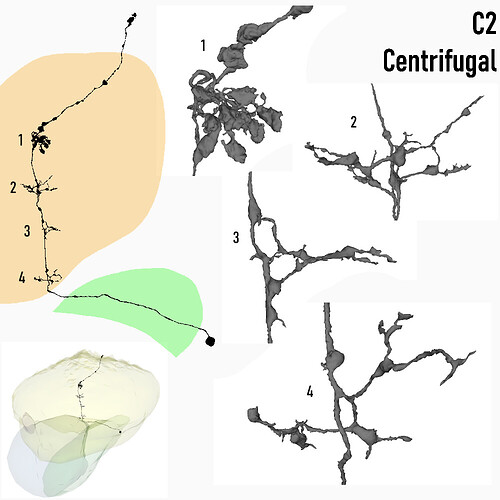
Centrifugal, C2
@AzureJay Yes, absolutely…all visualizations are helpful! It would also be beneficial to identify the cells in the text of the post, so that the post will be discoverable using the search tool, in the discussion board.
A list of keywords would work…for example,
Keywords: Centrifugal, C2
Nice visuals too, btw! ![]()
![]()
Cheers.
Keywords added for each image ![]()
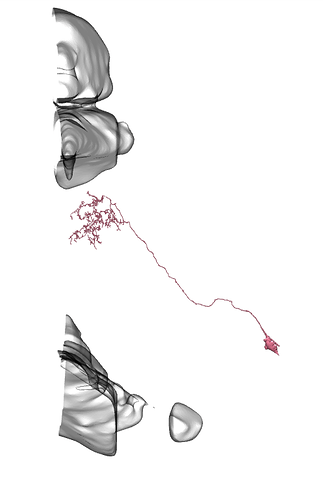
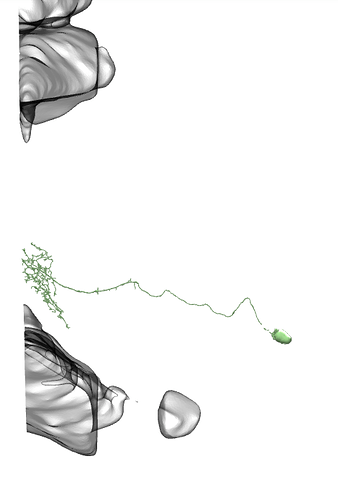
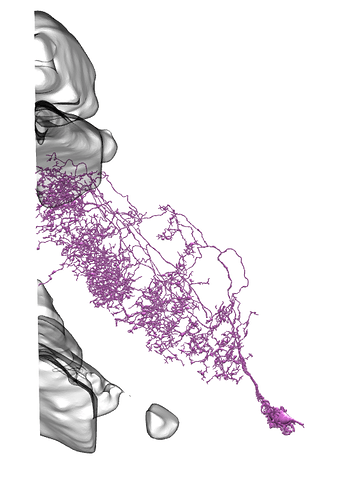
I found examples of Li11 - Li19 on neuPrintExplorer
You need to login with your Google Account to see the 3D renderings.
On the same site you can find more examples of the cells below (and of many others, ofc).
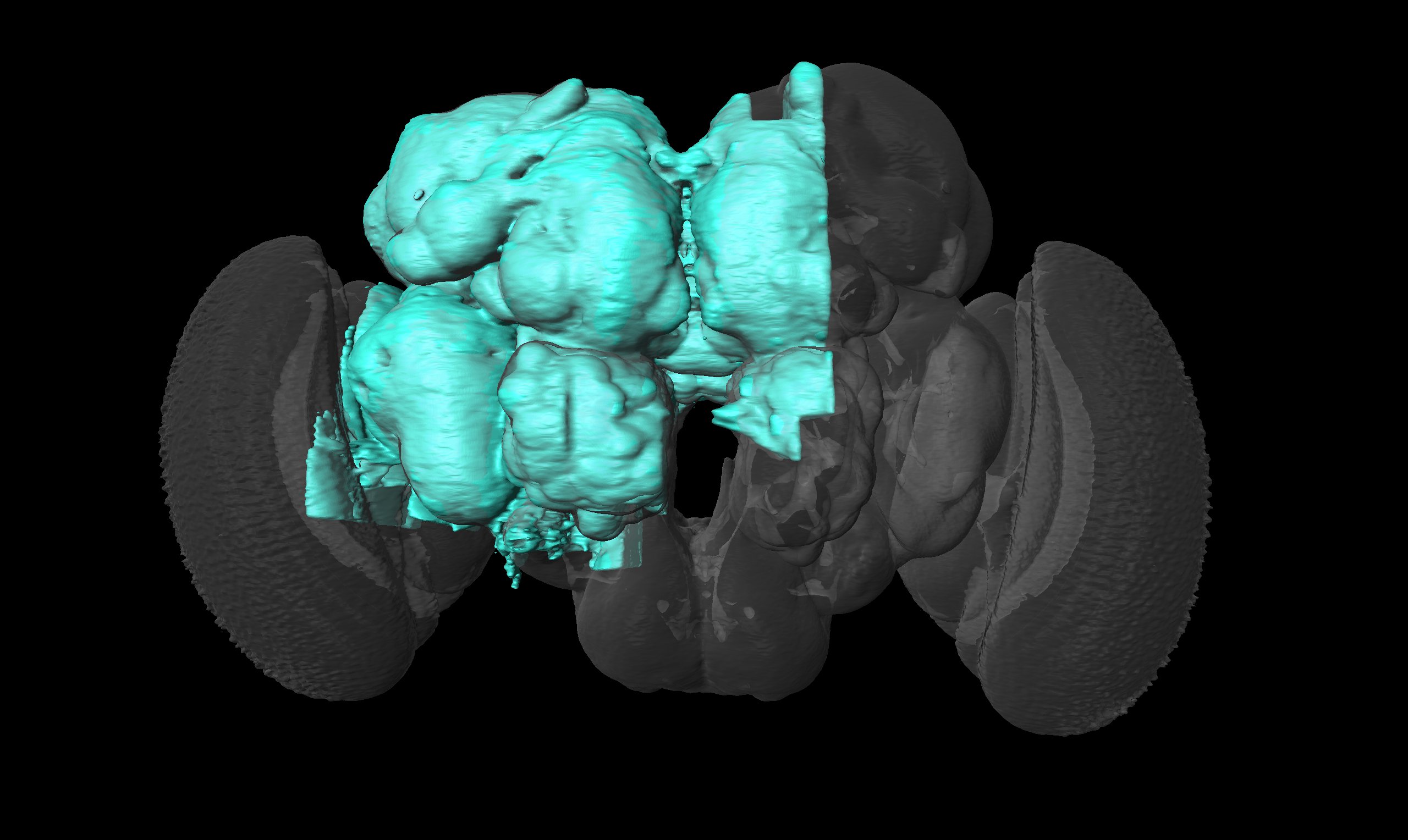
Unfortunately for us, they’ve focused on hemibrain, so not many cells from the Optic Lobe. They only have almost whole Lobula and some small parts of Medulla and Lobula Plate:
The 2 blobs at the top are parts of Medulla, the one at the bottom is LP, and the rest is Lobula.
Here is their whole dataset:
I didn’t find there any of Li1 - Li10.
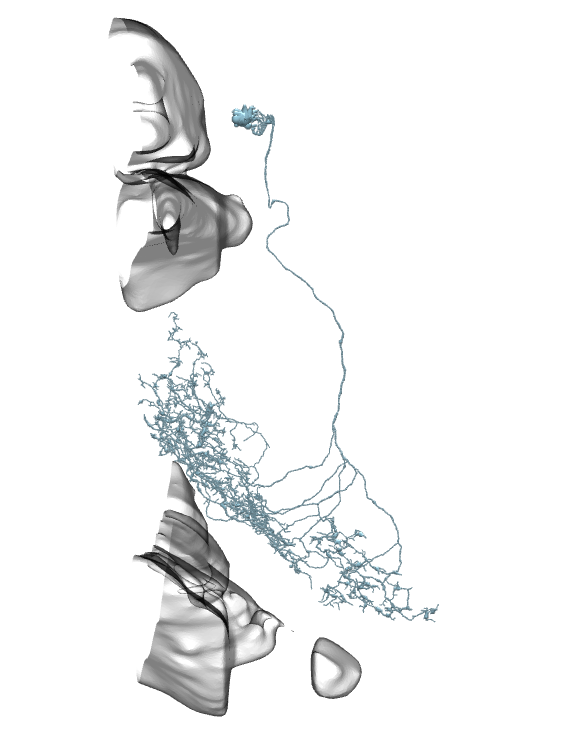
On all the images below I’ve added the fragments of Medulla (top) and LP (bottom) for easier orientation.
Li11:
Li12:
Li13:
Li14:
Li15:
Li16:
Li17: (this one has a big branch cut away, so be careful with identification)
Li18:
Li19:
I’ve also found there: Lt(1, 11, 33-38), Lpt(3, 4, 7, 8, 9) - they call them IVLPT, LPLC (1, 2, 4).
Keywords: Li11, Li12, Li13, Li14, Li15, Li16, Li17, Li18, Li19, Li20, Li21, Li22, Lt1, Lt11, Lt33, Lt34, Lt35, Lt36, Lt37, Lt38, Lpt3, Lpt4, Lpt7, Lpt8, Lpt9, LPLC1, LPLC2, LPLC4, lobula, lobula intrinsic, jamelia, neuprint
Edit: There are also over 30 000 cells identified that have input or outputs in Lobula (may take a while to load) - click the eye icon to view the skeleton:
Edit2: They have their dataset available in Neuroglancer, but it’s a different dataset from FAFB (as far, as I can tell), so the coords doesn’t much.
Their Neuroglancer:
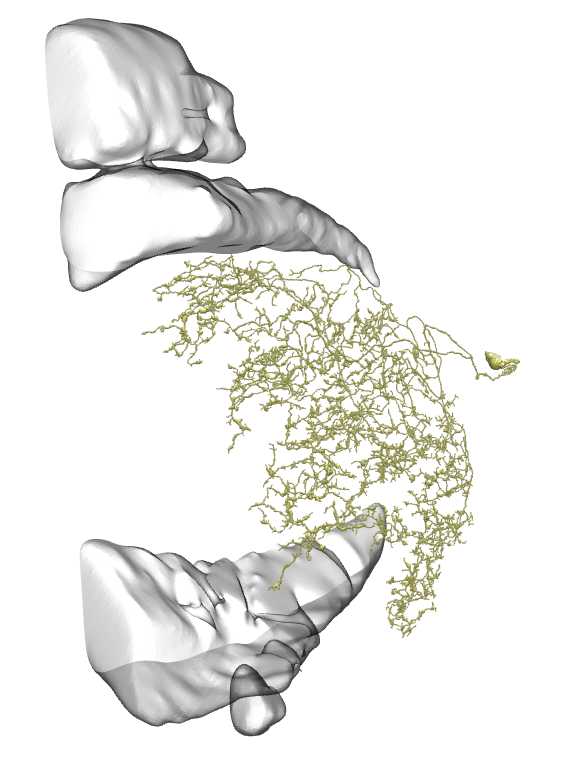
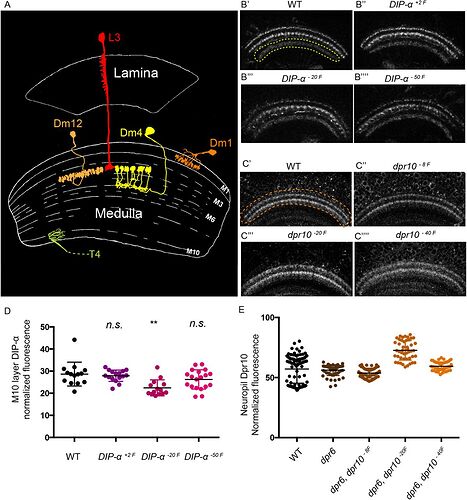
That looks more like dm cells than the same L cells we have from fishback, could ofc be a case of different naming?
The difference is, that Dm cells have their arborization completely inside Medulla and soma between Medulla and Lamina, while Li cells have arborization in Lobula and CB somewhere deeper in the brain.
And yes, these are all Li cells, not L cells. I’ve made a mistake and I’ll fix it very soon.
Keywords: L3, Dm1, Dm4, Dm12
What’s interesting in this picture is, that it shows Dm4 completely different than the Fischbach’s paper.
Other pictures in the Internet also show the Dm4 in the way, like the picture above:
So it looks, like the consensus amongst scientists is, that Fischbach’s Dm4 is wrong.
The Dm4 in Fischbach seems to only have one or two ‘clusters’ drawn, but the backbone structure, branch-twistiness, and location all look otherwise consistent to me. Maybe the full set of 20+ ‘clusters’ were omitted for “image clarity”? (Admittedly its the first time I’ve actually looked at the DmX section from Fischbach, I’ve relied on the images in this thread more for them)
Yes, the overall structure looks quite similar. The lack of the clusters confuses me. The clusters look like a defining feature of this type of cell.
Also, the one in Fischbach has mach wider arborization, that on the other drawings.
i have always looked at the Dm4 in fishback as someone having drawn it by hand, but not beeing able to adequatly represent the flower structure of a Dm4 so just making a crusedule instead.
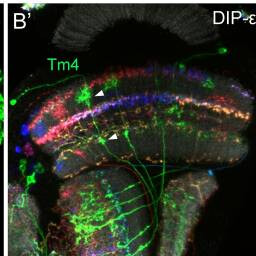
Tm4
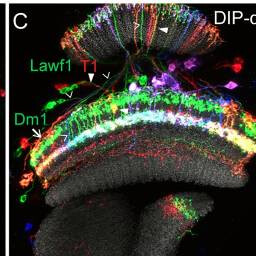
Lawf1, T1, Dm1
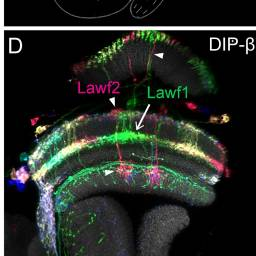
Lawf1, Lawf2
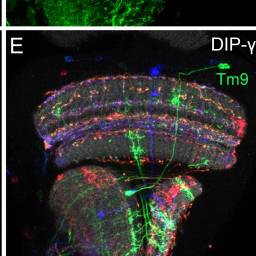
Tm9
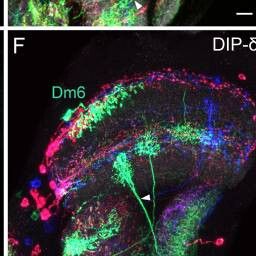
Dm6, Y3
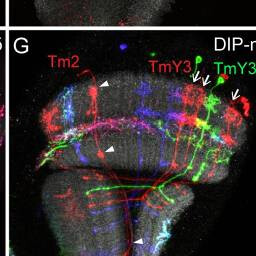
Tm2, TmY3
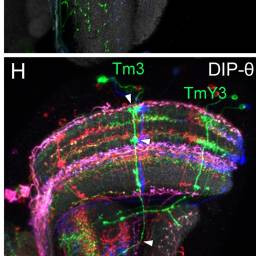
Tm3, TmY3
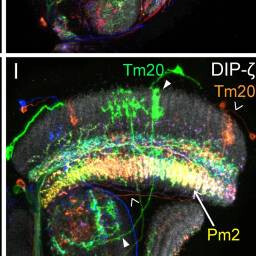
Tm20, Pm2
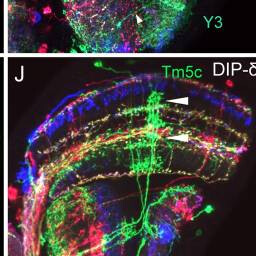
Tm5c
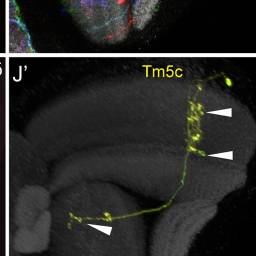
Tm5c
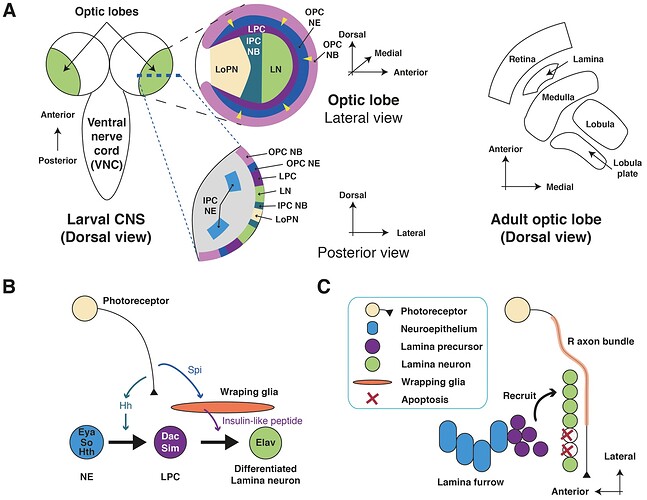
Note: I generally am avoiding any larval stage images to avoid confusion but sometimes they offer some interesting information. If this interests you I recommend Spatio-temporal pattern of neuronal differentiation in the Drosophila visual system: A user’s guide to the dynamic morphology of the developing optic lobe
Kathy T. Ngo, Ingrid Andrade, and Volker Hartenstein
Keywords: Optic lobe, Larval, Adult, Photoreceptor, Connectome, Lamina, Glia
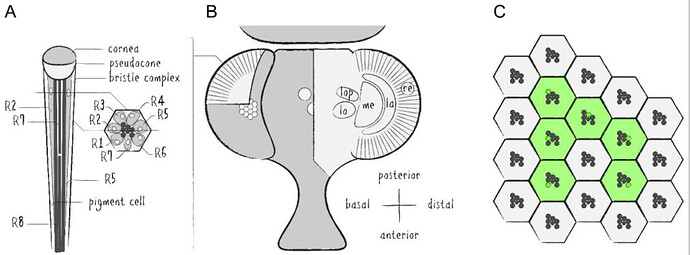
Schematics of the Drosophila visual system. (A ) Illustration of a longitudinal section through the ommatidial center (left side) and a cross section through the plane just below the pseudocone, where the nuclei (white) of the photoreceptor cells (PRCs) R1–7 (light grey) are located (right side). Trapezoidal layout of the outer PRCs’ rhabdomeres in the center of the ommatidium is indicated (dark grey). Primary and secondary pigment cells not drawn. (B ) Top view of a partially cross sectioned Drosophila head to illustrate the organization of the ommatidia (left side) and the optic lobe (right side). Indication of the hexagonal tiling by individual corneas on the curved surface of the compound eye and partial cross section to illustrate the wedged shape of ommatidial units. Order of neuropils from distal to basal: retina (re), lamina (la), medulla (me), lobula (lo) and lobula plate (lop). (C ) Illustration of the repetitive organization and identical orientation of ommatidia and PRCs in a top view of a section of the compound eye. Origin of the pseudopupil phenomenon is indicated by highlighting the fluorescence of individual rhabdomeres from adjacent ommatidia (green) which together generate the fluorescent ommatidial trapezoid when rhabdomeral FPs are expressed in R1–6.
Source: Application of Fluorescent Proteins for Functional Dissection of the Drosophila Visual System - PMC
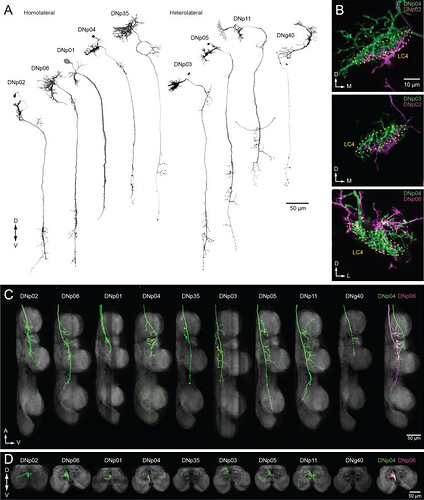
DNs forming a dendritic cluster within the LC4 glomerulus project to the lower tectulum.
(A) The morphologies of DNs which have dendritic innervation to the LC4 glomerulus (‘LC4 DNs’). The maximum intensity projection of a confocal stack with 20x objective are shown. All DNs partially share input (LC4 glomeruli) and most of them have axonal projection into the lower tectulum. These DNs are comparable to the ‘descending neuron cluster’ reported in blowflies (Milde and Strausfeld, 1990). (B) Simultaneous labeling of two different DNs innervating the LC4 glomerulus, visualized using multicolor flip out (see Materials and methods). Three examples are shown. The shape of the LC4 glomerulus is shown with a dotted line. (C) Sagittal view of LC4 DN axonal projections within the VNC. All but one (8/9) have axon terminals in the lower tectulum region of the VNC. One DN does not innervate this region (DNp03). An example of simultaneous labeling of 2 DNs is shown (DNp04 and p06, right). (D) Frontal view of LC4 DN projections in the mesothoracic neuropil. Projections are focused in the central region of the VNC volume, in the lower tectulum layer.
Source: The functional organization of descending sensory-motor pathways in Drosophila - PMC
Note: This source has a lot of great illustrations of the DNs (descending neurons); however we will likely not be encountering/working with them often. I chose this particular image since it interacted with LC4 and other LPLC neurons.
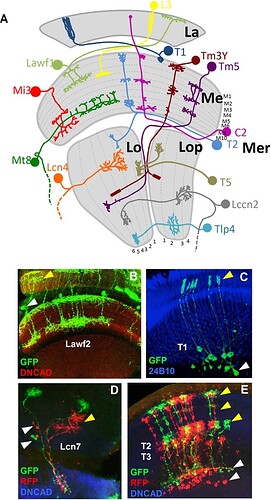
(A) Model showing a sample of neuron types in the optic lobe. The neuropils of the OL and the layers of each neuropil are shown in grey. La: lamina. Me: medulla. Lo: lobula. Lop: lobula plate. Mer: medulla rim. (B) Lawf2 neurons homogeneous MARCM clone. Arrowheads indicate projections in the lamina (yellow) and cell bodies in the medulla cortex (white). Projections reach M1 and M9 of the medulla neuropil (labeled with DN-cadherin in red). (C) T1 homogeneous MARCM clone. Arrowheads indicate projection in the lamina (yellow), cell bodies in the medulla cortex (white). (D) Lcn7 neurons homogeneous twin spot MARCM clone. Cell bodies (white arrowheads) and projections (yellow arrowheads) remain in the lobula. (E) T2-T3 neurons homogeneous twin spot MARCM clone. Projections are in the medulla (white arrowhead) and lobula (yellow arrowhead). (Neuropils in D and E stained in blue with DN-cadherin).
Samples summarizing the types of clones.
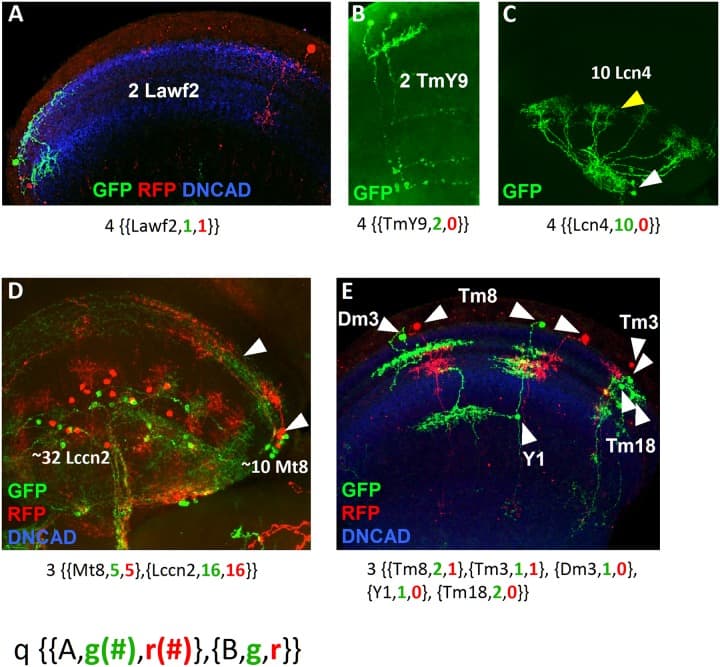
(A) Sister cell clones of Lawf2 labeled with GFP and RFP projecting in the medulla neuropil (blue). (B) Sister cell clone of TmY9 neurons labeled by GFP projecting in the medulla and the lobula neuropils. (C) Homogeneous clone of 10 Lcn4 lobula neurons, with cell bodies (white arrowheads) and projections (yellow arrowheads) in the lobula. (D) Cell clone showing two different neuron types: the lobula Lccn2 and the medulla Mt8 (white arrowheads point cell bodies in the medulla rim and projections in the medulla), both labeled by GFP and RFP. (E) Cell clone with 5 different medulla neuron types (Dm3, Tm8, Y1, Tm3 and Tm18) combining uni-columnar, multi-columnar, local and projecting neurons labeled by GFP and RFP. The nomenclature for each clone is indicated below. Medulla and lobula neuropils stained in blue with DN-cadherin.
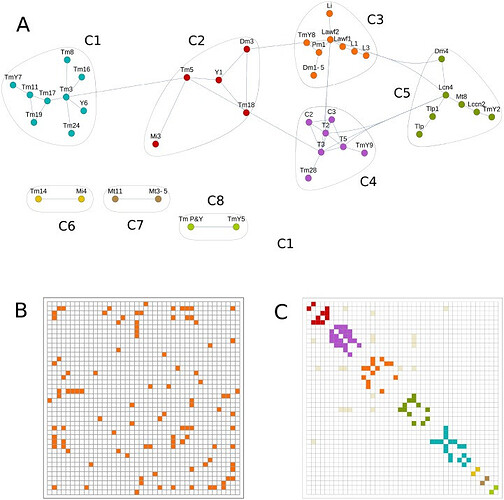
Community detection techniques find clusters of neurons related by lineage.
(A) Community detection graph for R≥0.75 and clones with quality of 2–4. This graph represents the optimal modularity value and identifies 8 communities (C1 to C8) of neuron types with tightly connected nodes, indicating that each group may be the product of one class of NB. (B) Adjacency matrix from the previous graph indicating the connected nodes. This matrix can be arranged in communities by adding a color code (C).
The above three images are from: A network approach to analyze neuronal lineage and layer innervation in the Drosophila optic lobes - PMC
Note: This offers some interesting looks at/discussion of ‘clone’ neurons and finding patterns in the macrostructures.