It’s good, but still missing many many cell types.
It’s possible to download the whole database to a .csv file and open it in Excel or any other Spreadsheet tool, but again, not all cells’ types are there. I’d say, about half, if not more optic lobe cells’ types don’t exist there. And some of the existing entries seems to be incorrect.
Also, I might be wrong, but I think, some cells can have multiple names and some names can be added to multiple types of cells depending, where in the brain a cell is.
yes i think DM cells is a example on this, there looks too be some marked as DM in the central brain and ours marked Dm in the optic lobe
@Amy_R_Sterling do you know what type of LPT neuron that is, in the fishback paper it is listed Lpt 1 and Lpt 2, but from what i understand there might be more?
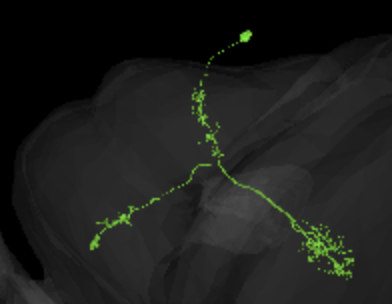
this is only marked as a y cell in the guide, i am thinking it might be a y6 any thoughs?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5989709234831360
For the DM versus Dm cells, it can be a bit confusing due to how the “m” is capitalized.
DM = Drosophila melanogaster (fruit fly)
Dm = distal medulla
And I’m not sure what type of LPT neuron it is (I’ll see if I can find any papers with more details). The Fischbach paper only has the “fragments of LPT 1 & 2” in it and the writers concede that their “list of tangential neurons in the lobula plate of Drosophila has to be extended” so there are more types out there…
I think, it looks more like a Y3, because of how deep it penetrates the lobula. Y6 would be my second guess.
yes, it goes a bit deep, the reason i tought it was Y6 was because that extra twig before the main cluster in lobula on the Y3 cell
@annkri I come up with the same as @Krzysztof_Kruk …it looks like a toss up between Y3 and Y6.
Also, I don’t see any reference to Y2 in Fischbach…is there a Y2? If so, I’m curious what it looks like.
yes, I see the branch and wonder, if it’s a defining feature for Y3 or just something, that might happen to particular examples of them, but not to all of them.
I found these descriptions for both kinds:
Y3:
Wide-field Y neuron with a mix of bleb-type and fine arborizations in the lobula plate. These show some stratification, but cover all layers to some extent. The medulla branch has a relatively wide, terminal arborization field in M8-10 with a mix of bleb-type and fine arborization. The lobula innervating branch has narrow arborizations, also of mixed type in layers 2-5 (Morante and Desplan, 2008; Fischbach and Dittrich, 1989)
Y6:
Y neuron that arborizes relatively broadly in all lobula plate layers and with a mix of terminal morphologies. Its branch in the lobula has narrow and mainly bleb-type arborizations in layers 1-4. Its branch in the lamina has a broader, bushy, fine, terminal arborization domain in layers M8-9.
Still, hard to say.
@TR77 Yeah, indeed Fischbach doesn’t have an Y2.
@Krzysztof_Kruk Oh good, then we can just define anything we’re not sure about as “Y2”, lol. ![]()
On a more serious note…
Now that HQ has given us the go ahead to start identifying cells in the database, should we take a moment to agree on a naming convention, for identifying cells, so that our entries are consistent in the database? Any thoughts?
Out from that description it sounds very close, but also like two different people has written it so very difficult to make a decision based on these descriptions, in particular when only knowing half the yargon.
Oh, didn’t read the post yet, thanks.
As for having a consistent naming convention, I totally agree. For now, I’ve only named a couple of Mi cells, and wrote “Mi1” or something like this, and one Am/Lai, where I’ve given a source.
Agree. It would be cool, if we could somehow recognize, where the internal layers in 2D and 3D are in all 4 components of the optic lobe.
i have noticed that in most of the neurons in the optic lobe they write things like TmY15, TmY15_R is this showing if it is in the right or left lobe (what is the front if that is the case)
I was about to ask the same thing…should we add a note to distinguish between the left and right lobes?
A shorthand notation for identifying cells sounds good to me (e.g. “L1”, “Mi9”, “Dm4”, etc.), if no one has any objections, or anything else to add.
@annkri Just a friendly fyi…
As I was looking for more examples of Tm1a in our dataset, I came across this reference in Fischbach, that explicitly distinguishes between Tm1 and Tm1a,
“It is instructive to compare variants of the same type, e.g. Tm1 with Tm1a, and Tm18 with Tm18a. In these cases the arborizations in the proximal medulla either sprout directly from the main fiber or they are connected to it by a stalk.” (Fig. 8, Fischbach)
Looking at the diagram in Fig. 8, you can indeed see that Tm1a is shown having an arbor in the lower medulla, that is slightly separated from the main backbone, by a “stalk” (as the paper calls it).
Again, just a friendly fyi…
Cheers. ![]()
The few id’s i’ve put in so far have been a copy-paste of the researchers identifications (in my case Dm4_R;Dm4;) - for left vs right, here is a link with a Dm4 from each side identified by not-me ![]()
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6437728178667520
@st0ck53y Okay, I like that…
I’m not sure what the benefit is in using the redundant id separated by semicolons (i.e. id_L;id). It seems like we could just list the id once, maybe (i.e. id_L)?
Also, just so we’re clear and all on the same page…using the above convention, our optic lobe is the Left optic lobe, correct?
Yeah, i think most (if not all) of the assigned cells at the moment are in the left lobe. not sure how i got over to the right for my current lot ![]()
As for the redundency, the only thing i can think of would be for some kind of exact-match querying, but no idea tbh. The ones ive checked had it, so im just doing the same for now
@st0ck53y Got it…perhaps it would be worthwhile to run this by HQ real quick. @Amy_R_Sterling @M_Sorek
Thank you for finding the official difference betwheen the Tm1 and Tm1a