Yes you’re correct, it’s a new dataset. Here is the chart
The generation of a whole larval zebrafish brain electron microscopy volume in tandem with automated tools lays the groundwork for producing the first vertebrate brain connectome.
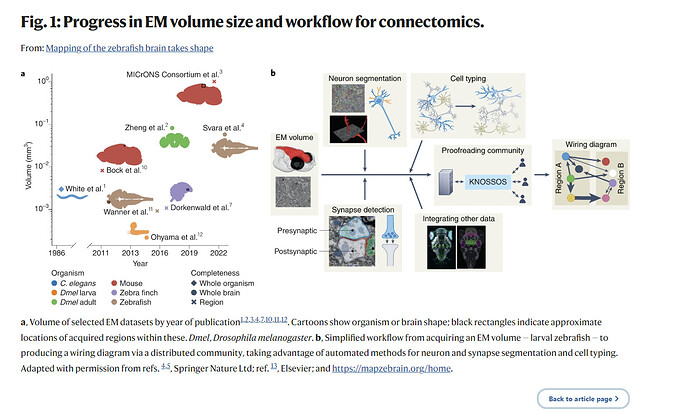
Understanding the relationship between behaviors and the circuits that generate them is a major goal of neuroscience. Connectomes or static wiring diagrams of the nervous system provide a basis for generating testable hypotheses on neuron function, contributing appreciably toward this goal. Since the first connectome, that of Caenorhabditis elegans with 302 neurons1, was published in 1986, the field has advanced slowly, but the pace has picked up recently. In the past 15 years, substantial progress in methods and technology, from sample processing to neuron reconstruction, has allowed a gradual increase in volume size, density of reconstruction and neuron completeness for a variety of vertebrate and invertebrate species (Fig. 1a). In particular, improvements to automated methods for segmenting crucial features from the image data, such as neurons or synapses, have led directly to a notable decrease in reconstruction time by providing a more complete starting point. Volumes of the size of whole brains for the adult Drosophila melanogaster are now achievable2, allowing comprehensive mapping and identification of most neurons in those volumes. There are, however, still substantial constraints on the attainable dataset sizes. In vertebrates, the biggest volume to date has been the recent MICrONS data set comprising 1 mm3 of mouse visual cortex3. Now, Svara et al.4 generate the first electron microscopy (EM) volume of a whole vertebrate brain (excluding the retinas) for the larval zebrafish (Danio rerio) at a resolution sufficient to map a complete wiring diagram. By applying previously established and new methods developed by Schubert et al.5 and putting in place the required infrastructure, the potential for the community to generate a full larval zebrafish brain connectome is now within reach.
Entire article:
Recent developments in connectomics have highlighted that the time and resources needed for neuron proofreading are currently the limiting factor in generating dense connectomes.
Bigger volumes might therefore be less densely reconstructed, substantially affecting their usefulness to their fields. One way to overcome this is to decouple volume generation from proofreading, providing the infrastructure for a decentralized proofreading community, composed of interested research groups, to form. Putting this into place, however, also demands resources, community management overheads and a strong commitment to open research. Another way to add value and biological relevance to a connectome is to link published data such as gene expression, transgenes, neuron function or neurotransmitter identity to reconstructed neurons. This usually happens at the level of cell types, not individual neurons. This integration provides a much richer landscape with which to interpret the circuits and the tools to test them (Fig. 1b).
Svara et al.4 present an impressive advance for the zebrafish community, as until now only subvolumes of the larval brain had been acquired and reconstructed. The volume encompasses 0.058 mm3 at a resolution of 14 × 14 × 25 nm3, which is sufficient for synapse detection, and contains an estimated 121,000 neurons. Svara et al. take advantage of automated methods, and in particular those reported by Schubert et al.5, to identify synapses with the help of the SyConn2 toolkit. By reconstructing more than 400 partners for two interneurons involved in processing visual stimuli, Svara et al. thus demonstrate that circuit mapping is possible and can be done very efficiently in this volume.
Individual neurons imaged by volume EM (vEM) require meticulous reconstruction. This once required skilled researchers to manually trace each neuron, with Svara et al.4 reporting around 60 h to complete a single cell. Manual reconstruction is therefore a substantial bottleneck for gathering large-scale connectomic data. By contrast, applying and tuning a recent machine learning algorithm to segment neurons from vEM datasets6 substantially reduces the time needed to reconstruct each neuron to 1 h and replaces up to 94% of the human interaction. With 121,000 neurons to proofread, the automated approach considerably reduces the theoretical estimate of 7 million human hours to annotate the entire zebrafish brain to a still substantial 5,000 days, or 13.8 years. Driven by the aim to make their EM dataset and the necessary proofreading accessible to the zebrafish community, Svara et al. extend the Knossos 3D proofreading tool to support a live environment and dozens of concurrent users. A similar community approach to generate a fly brain connectome has, after a few years, over 200 users from 50 labs7. Engaging the zebrafish researchers and providing intuitive and robust tools for proofreading and analysis will be key to developing an active community and producing a good-quality and comprehensive brain connectome in the near future.
Svara et al. also map their connectomics data, via image registration, to functionally classified optic-flow neurons and to the zebrafish atlas Mapzebrain (mapZebrain), which includes light microscopy-level images for transgenes, brain region annotations and single neuron morphologies. This integration will be essential for testing neuron functions inferred from insights obtained by connectomic analysis.
Quantifying how the shapes of neurons, synapses, and other intracellular structures vary adds information to the connectivity wiring diagram, which aids the analysis and identification of neuron types. Schubert et al.5 present a new method that can potentially automatically classify neurons into types. For each neuron, their algorithm extracts a set of features describing these segmented structures. The similarity of these per-neuron feature sets, called embeddings, clusters populations of neurons into putative cell types without the need for prior hypotheses about types or time-intensive annotation of types.
Application to an area of basal ganglia in the brain of the zebra finch demonstrates this potential. Without expert supervision, SyConn2 discovers clusters of neurons in the embeddings consistent with previously known types. Analysis of the segmentation reveals correlations between differences in cell types, mitochondria–synapse distance distributions and firing properties of those types, consistent with observations in prior work. Such functional correlates latent in vEM datasets become discoverable through tools such as SyConn2 that describe aggregate structural properties across whole neurons and cell types.
SyConn2 improves on similar methods8 by representing segmented structures as sparse points sampled along their surface. Embeddings are learned for these points using recent developments in point cloud processing with deep neural networks9, which is shown to computationally scale linearly with the size of vEM datasets, along with the rest of the segmentation and processing. SyConn2 is open-source and built on cloud infrastructure so that it can be reused by other researchers and scaled to resources proportionate to each dataset. Further iterations of the method should test its generality, as well as demonstrating that the classifications obtained are comprehensive and able to reflect the highest cell type resolution as defined by human annotation.
While unsupervised methods like SyConn2’s neuron embedding reduce the requirement for human-generated annotations and may reduce bias in downstream semantic analyses such as cell typing, most time costs remain concentrated in the annotation and proofreading required for segmentation of neurons and synapses. Scaling connectomic software tools to larger volumes requires simultaneous progress on many fronts. Improvements at later processing stages can feed new data, insights, and constraints into earlier stages. For example, detecting that features of part of a neuron are inconsistent with those expected by its type can be used to correct merge errors in segmentation.
The rapid progress seen in connectomics in the past 15 years has started to bear fruit, with whole brain connectomes now possible for invertebrates and vertebrates. The work described by Svara et al. and Schubert et al. in this issue is a major step, as the researchers not only generated an EM volume, but also provided the community with a resource and tools to engage with and benefit from the connectome reconstruction efforts. Until automated tools are further optimized, community involvement will be key to generating full and complete connectomes.