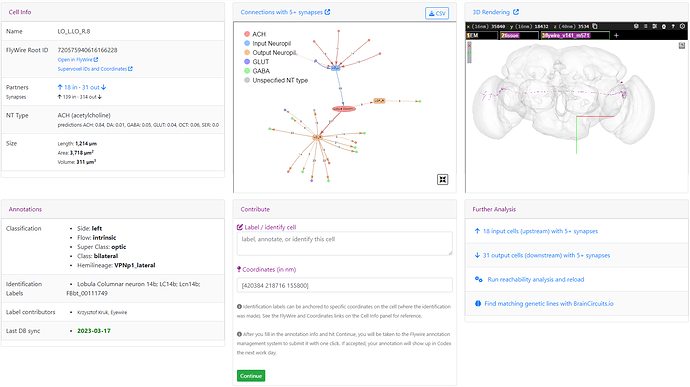
v 0.2
Fixed an issue with copying visible segments.
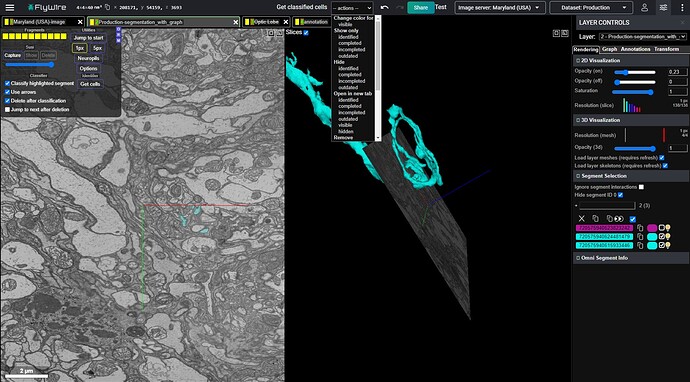
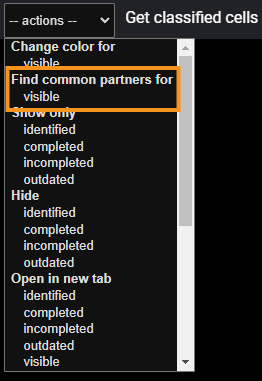
Added a large feature to find common synaptic partners for up to 10 selected cells and to find further synaptic partners of those common ones.
Warning: There are still many issues, that have to be ironed out, especially with the second part (finding partners for partners), but it’s already usable, that’s why I’m publishing it.
The first step to use this feature is to select up to 10 cells in the right panel. If you select more, the first 10 will be automically selected.
After that, click the appropriate option from the dropdown menu at the top:
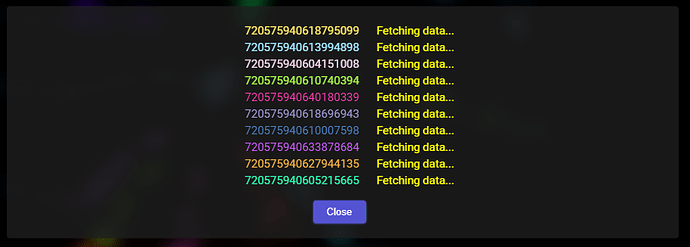
At the beginning you should see a dialog window similar to this one:
The addon is now retrieving all the necessary data from the Connectivity app’s API.
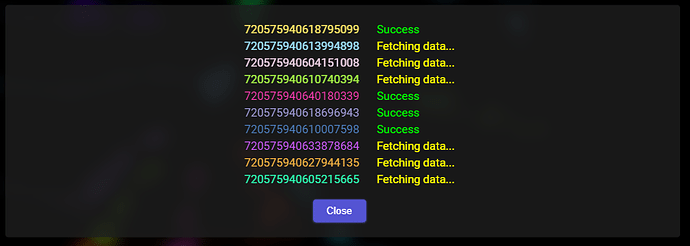
With time, whenever the downloading of the data for a given segment is successful, the window should look like this:
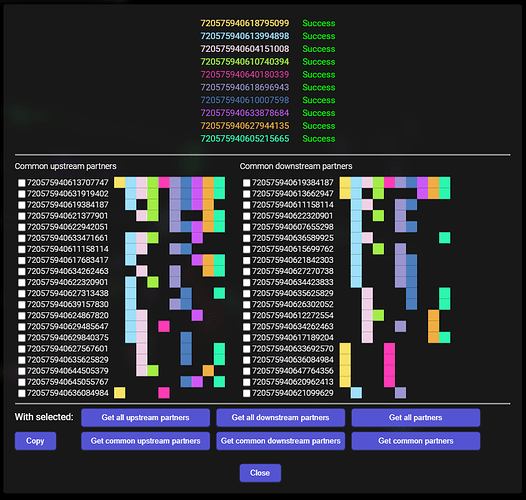
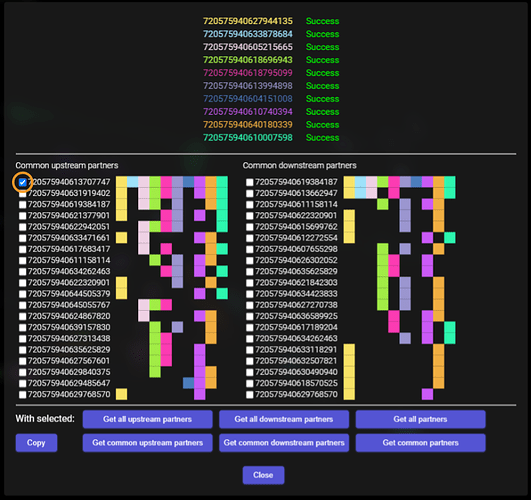
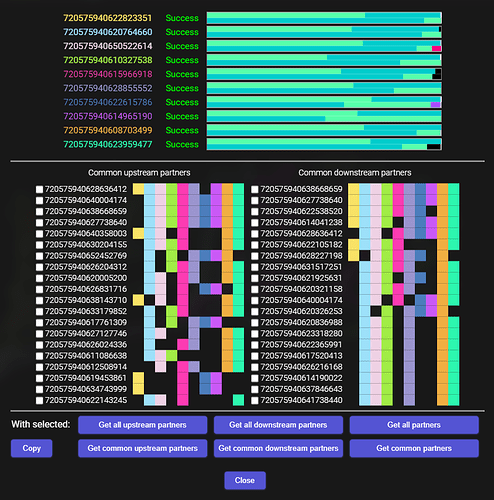
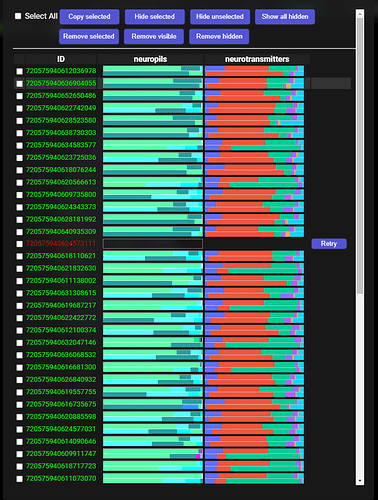
When everything is downloaded, the addon does some extraction and grouping of the data and should present the results in this way:
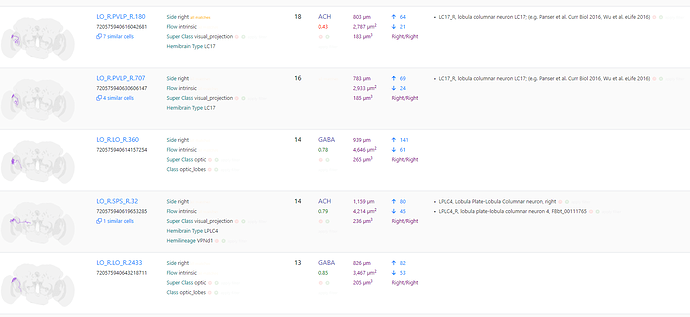
The segments at the top are, of course, the ones, with which we started.
Below that are two columns - the one on the left is a column containing first 20 segments that are most common upstream synaptic partners to all the starting cells. By most common, I mean, synapses with the most of the starting cells. The column on the right is the same, but for the downstream synaptic partners.
The squares to the right symbolize the starting cells - each color is the same, as the color of the cell’s ID at the top. So, for example, the first common upstream synaptic partner has a colored square for each of the starting cells, which means, it sypanses with all of them. The next one doesn’t have any connection with the first and the fifth starting cell, and so on. Same for the downstream partners.
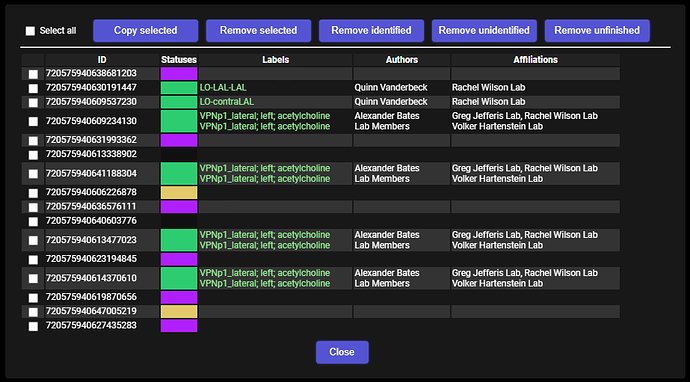
At this moment you can finish using the tool - select checkboxes to the left of each interesting cell and click the “Copy” button to copy all the IDs to the clipboard.
But you can do some further work with the other six buttons at the bottom.
This time you can find various synaptic partners for the newly found cells.
To do this, mark the appropriate checkboxes and click one of the buttons below.
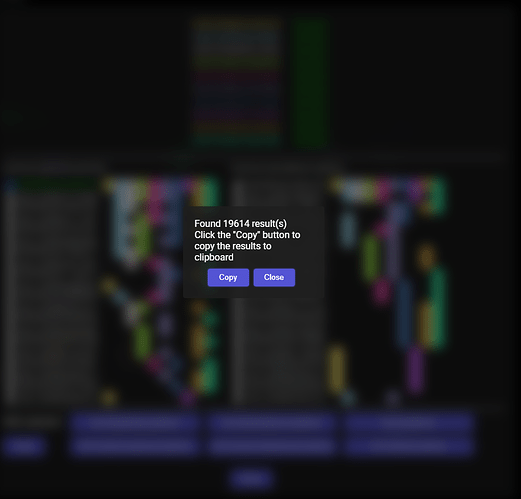
The first row of the buttons will find all the synaptic partners for all the selected cells (all upstream, all downstream, or both). It’ll remove duplicate entries, sort the list in ascending order and show a popup with the number of found cells. You have to click the “Copy” button in that popup and all the IDs will be copied to the clipboard (comma separated list).
The second row is for searching common synaptic partners (again, upstream, downstream or both). “Common” means, that each given ID on the final list is a(n) (upstream, downstream, any) synaptic partner to all of the selected cells. In other words, it’s an intersection of all the sets, where sets are the lists of synaptic partners of the selected cells.
After you’ve selected all the needed cells and pressed an appropriate button, the selected cells’ IDs should turn yellow. It means, the data is being pending/fetched for such cell. If a cell turns green, it means, the data has been completedy downloaded for it. If it turns red, that means, that something went wrong with downloading the data.
When all the cells turn green, you should see a popup informing you, how many synaptic partners has been found. There’ll be a “Copy” button to copy all the IDs to the clipboard.
-
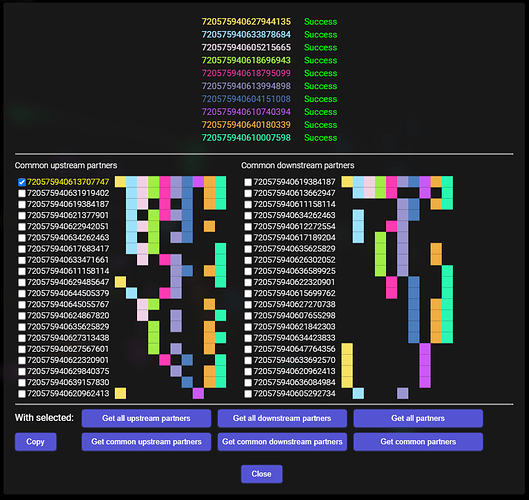
Selected one of the found synaptic partners:
-
Addon during downloading the data for the selected partner:
-
Results found after clicking the “Get all partners”:
Collecting the data at this point might be quite long, because the common cells are usually large ones.The longest time, I had to wait, was over 4 minutes. It was for cell 720575940619384187. And it’s one of the biggest, most complicated cells, I’ve ever seen. It is composed of 8346 chunks, the file with synapses is 24,5MB in size and the segment have already had 5664 changes.
Remember, that the final results are for the selected cells, not for the starting ones.
So, there are 5 levels of connectivity, that you are able to reach (2 upstream, 2 downstream, starting one):
- starting cells → upstream partners (1 level up from starting cells) → their upstream partners (2 levels up from starting cells)
- starting cells → upstream partners → their downstream partners (same level as starting cells)
- starting cells → downstream partners → their upstream partners (same level as starting cells)
- starting cells → downstream partners (1 level down from starting cells) → their downstream partners (2 levels down form starting cells).
Why did I do this feature?
To find as many as possible cells of the same type.
For example, I can start with 5 Mi1 cells. Then I’ll search for their common upstream partners and for the most common ones, I’ll seach for their common (or all) downstream partners. (or downstream and then upstream). This way, I’ll end up with not only the Mi1 cells, I started with, but probably with most (if not all) the other Mi1 cells (among many other cells, but it’s much easier to filter out, that to search via random clicking on the 2D).
Issues/limitations:
- Not all errors are currently catched, so some may silently raise in background without informing about it.
- When searching for synaptic partners of multiple selected cells (from the second part), the highlighting doesn’t always work correct.
- It’s best not to select more than 20 of the synaptic partners, because there are some limits in browser about max number of concurent downloads.
- If you had four or more errors (red cells’ IDs), the browser might block you for 15 minutes for further reaching the Connectivity app via the addon. You can read more here: Anti-DDoS HTTP Throttling of Extension-Originated Requests