Thanks for the info. However, this link is righ-click-copy-pasted from the main link, so I’m not sure, what could possibly be wrong ![]()
![]()
![]() hahaha
hahaha ![]()
Would it be possible to some how copy the IDs of the cells that got an error during submit phase, or the ability to highlight the text of the ID so one could copy it? Sometimes the lightbulbs don’t change color and I want to make sure I labeled all of them while lightbulb blind.
v 0.3
Added a counter showing, how many entries there currently are.
Added a button to copy IDs of all the currently visible entries.
Added backtick (`) as another shortcut for marking cells.
Added checking for correctness, if the cell added to the list is indeed the one, we wanted it to be. To make a correct addition, you have to position the 3-axis cursor inside the cell (as it was before) and hover the mouse cursor over the cell, you’re submitting. I doesn’t have to be hovered at the exact moment, you’re submitting the cell, but it has to be the last cell being hovered before the submitting.
The checking works by comparing the ID gotten from the 3-axis cursor with the ID gotten from the last hovered cell. If the IDs differ, the small square in the left top corner will turn red.
Edit:
v 0.3.1
Added “Copy by label” functionality. After clicking the button in the top part of the “Get cells” dialog there will be shown another dialog, which will list all labels and all IDs assigned to each label. A button in each row will copy the IDs for that given label to the clipboard.
Hey I was using the identifier today when I noticed that I’m getting red boxed when I try to add cells to the list for batch labeling and they don’t show up on the list. Some of these cells might been previously added to a classification, if that changes anything. Any guess on how to proceed?
Make sure, that the 3-axis cursor is inside the cell (not on its surface) and the mouse cursor is hovering over the same cell.
It still won’t add the cells to the list.
Ahh ok it works if I hover over in the 3D. Is there any way to turn off this safety?
Also weirdly won’t work for all the cells.
Currently, there isn’t a way to turn it off. I’ve added it because in about 1% of identifications there were errors.
When I’m marking cells, I close the 2D panel completely. That’s why I didn’t think about hovering over cells in that panel.
If it doesn’t work for a cell, try changing the position of the 3-axis cursor and make sure, that the cell is up to date. It sometimes happen, that the 3-axis cursor is placed inside some small fragment, that isn’t merged with the main cell and that leads to IDs being different (and incorrect labeling).
Ok I’ll test out some different positions. I don’t mind the 1% error rate for the massive speed increase in the short term, although I can see 1% being bad in the long term. That being said I think anything with more than one label will be checked in the future. For us currently, we are trying to label as fast as possible before the end of the month. Having to maneuver the cursor and the crosshair and then not being able to know which of the 20 cells didn’t get on the list is gonna be a big road block unfortunately. I might have to switch back to manual labeling simply for speed.
When I was testing this solution, I haven’t had any significant decrease in the speed of labeling; 10% at max. Considering, that I’m able to mark 700 cells in an hour (when I have them already checked earlier), 10% difference is just 6 minutes. And even then, I think, it’s still much faster than manual labeling, when one have to have the 2D panel opened, wait for the 2D chunks to be loaded, then click 2 times, paste the label, click again, wait for the submission and close the new tab; everything in series.
However, if you want the script without the new features, here’s that version:
Just edit the file in TamperMonkey, remove the lines near the top, where it says:
// @updateURL https://raw.githubusercontent.com/ChrisRaven/FlyWire-Cell-Identification-Helper/main/Cell-identification-helper.user.js
// @downloadURL https://raw.githubusercontent.com/ChrisRaven/FlyWire-cell-Identification-Helper/main/Cell-identification-helper.user.js
and save the changes. This should prevent automatic updates.
Awesome thank you! I’d be interested to see your method for labeling. Currently I paste a list of candidates into splitter and go 20/20 removing incorrect matches and labeling the rest. Do yall have a better process that might help? If a cell is black lightbulb then it can’t be labeled? We are taking cell IDs from codex and if we refresh the ID to label them will that still show up in codex later I hope.
I’m doing the whole proces in stages.
I start with all the completed cells and using the splitter, I’m looking for all cells of one single type. Using the Classifier, I’m adding all the correct one to the list. When I have most of them, I’m using the Batch Processor to find synaptic partners of the most common synaptic partners for the already found cells an then do the Splitter thing again, for that new list.
When I have all (?) cells of the same type, I’m using the Splitter again, just to be sure, that all the cells are indeed of the same type. During this process I’m also fixing some small issues, like missing parts or clearly visible mergers. This time, I’m using the “Save left” functionality to collect all the correct ones.
Then I check for the outdated ones using the features from the Batch Processor (open outdated in a new tab and remove outdated from the current tab) and update them.
Finally, when I have all the cells done, I’m using the Splitter yet again, this time for labeling only.
This whole process can take a whole day or even more, but it helps with finding all the cells of a given time and helps with identifying ones near the edges of a neruopil, because when I’m working with a single type, it’s easier to recognize even the malformed cells.
With the older version of the addon it was possible to label the outdated (black lighbulb) cells. However, that can lead to some issues, if the updated version turns out to be a different type.
The updated cells should show in Codex.
Updated cell IDs with labels will show up when Codex’s version is next updated.
For example, this is why we are missing so many labeled photoreceptors (R-cells) in Codex right now. The photoreceptor segment id’s weren’t “completed” when the current version of Codex was updated (version 630). Only completed cells are listed on Codex. So the R-cells are completed and labeled in FlyWire’s database, but that is separate from the versioned Codex db.
There’s also a way to get all the completed cells up to any date using the “Cell Completion Details” tool, but I’m not sure, if I should share the method here, on the public forum.
I am running into some sort of error when trying to mark cells but I can’t pinpoint what’s causing it.
When I go to mark most cells I am getting a red error box instead of the green one, and it doesn’t add to the identifier list.
For example, when trying to mark (68362, 58004, 6697) coordinates, which I can verify in 2D and 3D are part of the cell data, I get a red square. Moving to another coordinate does not fix this.
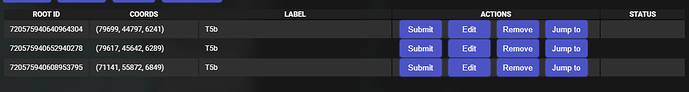
The coordinates above belong to root 720575940617509947, and you can see here it is not part of my current list:
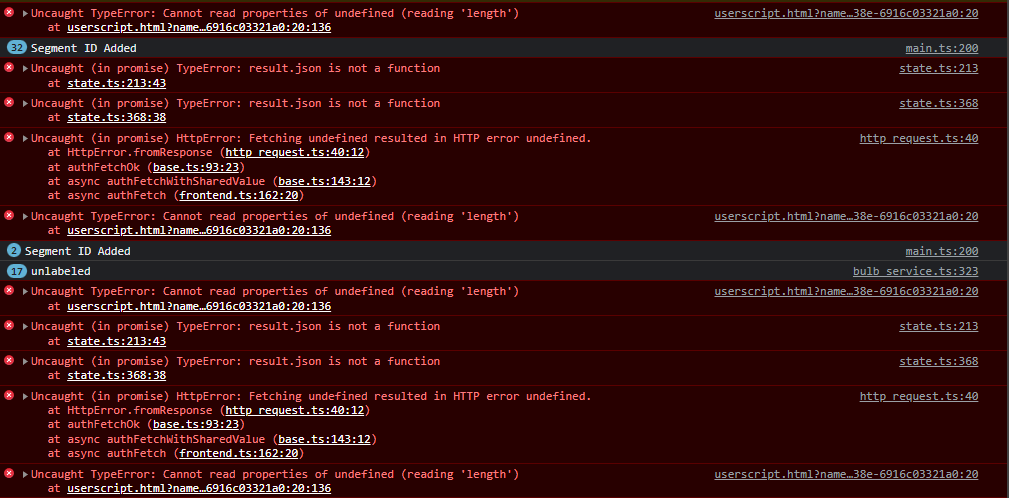
Here’s what I was able to read from the console (Chromium based browser, Vivaldi) -
Happy to try to offer more info if I can - this didn’t happen until this newest version ( 0.3.1).
I think, I know, where the issue is. You probably have the neuropils turned on. If you hover your mouse over any of the neuropils, it’s still treated as a cell and collects the neuropil’s ID, then compares the ID to the one of your segment. The comparison fails and the red square appears. At least, that’s what happened to me, while testing this problem.
For now, I don’t know, how to solve this issue other than:
- turning off the neuropils,
- hovering the mouse over the cell and not the neuropil, or
- removing this functionality from the addon (by me, in future versions, ofc).
Some other co-workers were wondering how you do this part since they deal with a lot of outdated cells sometimes. I was able to figure out how to get them to a new tab, but I’m stuck on the part where you update them in batch. Or do you still deselect and reselect one by one?
Unfortunately, I do them one by one. I was trying do do it programatically, but haven’t had much luck. So I just set the slides resolution to low (5px) zoom in to 300nm, so the slides load as fast as possible and do the clicking.