let us do thiiiiiiiiiiiiiiiiiiiiiiiiiissssss!! lol
@Nseraf Yeah, clearly a MASSIVE parallel processing layer (i.e. Photo-receptor > Retina > Laminar Cartridge > Medulla > … > Central Brain). It reminds me of things I have seen in industry.
@Krzysztof_Kruk I was thinking something along the same lines, as well. Rather than creating a guide specific to laminar cells, I would like to ultimately create a general guide to inferring microstructure from macrostructure (when the need arises, due to problems encountered in the EM data). I’m picturing a collaborative wiki-style document, where we describe useful macrostructural patterns, as we encounter them, and characterize the associated cell types (including useful recognition heuristics, such as the L5 “Deer Antlers” that I pointed out above).
At the moment, I would like to extend this concept of inferring microstructure from macrostructure, based on structural regularity, to the Medulla, and then on to the Lobula areas. If anyone has any observations that would be helpful, feedback would be most welcome.
Cheers, all.
Just wanted to note here…
Thanks @annkri for putting together a cell identification sheet, in the FlyWire Q&A Log!
Cheers.
When I was browsing the identified cells, I’ve noticed, that some of them are missing some structures.
Link: Sign in - Google Accounts
You can type “missing”, “lacking” or some other helpful keywords to find those cells.
I’m wondering, if we could find the missing parts. In some cases, it would be, of course, impossible, like in cases, where something is missing because of the dataset edge or missing segmentation, but some entries saying “missing branches” sound, like something could be done with it.
Keep in mind, some of them are in older segmentation, so you’ll have to click the red clock icon on the “Production segmentation” tab to update it.
Not sure, if it would help and if we should update the identification descrption after we find something. So it’s a post just saying, that such things exist.
EDIT: we could use that page to find cells of types missing in the spreadsheet
I’m not sure we can edit those cells, I tried to add an extension to the last one on the link and it tells me “merge does not work with a segmentation at an older state” and the same happened when I tried to remove a merger, “split does not work with a segmentation of an older state”.
But having said that, even if we can’t edit them, maybe if one of us does find a valid connection between one of those cells and the cells in the q/a sheet an admin can merge them together and complete the cell(s).
Did you try to update the segmentation?
other than double click both in/out is there a diff. way?
By clicking on the red clock icon on the “Production segmentation” tab.
ok i did not know that, hah. Never had an instance requiring it before. Thnx!
If anybody wants to see parts of the Drosophila brain, you can open new tab (inside FW) and paste this link
precomputed://gs://flywire_neuropil_meshes/neuropils/neuropil_mesh_v141.surf_v2
into the source field, then click inside the Name field and then save.
After that you can switch to the new tab and add fragments from 1 to 74 to see various parts of the brain in different colors.
You can change the 3D opacity of this layer and combine it with some cells to get some nice pictures.
Fragments, you may be more interested in, are: 2, 6, 14, 29, 35, 43, 51, 56 (parts of the optic lobes).
@Krzysztof_Kruk Are you by any chance familiar with the syntax for querying that database (identified cells)? Any tips for us? It looked to me like there is at least SOME inconsistency in the naming of identified cells, in that database (haven’t actually verified that yet, though).
Oh and, awesome tip about the neuropil meshes…thanks!
Unfortunately, I don’t know the syntax. I’m just entering things, I’m looking for in the “by” field, when the “Filter” is set to “Cell identification” and get some results. It searches by all fields, so the results might be polluted sometimes. However, sometimes, it can return interesting results, like this one, when I’ve entered “Lab” in the searching field:
And, yw! ![]()
Edit: well, actually, it searches only by the “Cell Identification” field.
Edit2: when you don’t enter any search term, just click on the top link named “Cell identification” it returns all identified cells (over 60k already!):
https://prod.flywire-daf.com/neurons/api/v1/cell_identification
@Krzysztof_Kruk Ah, I see…I was hoping to narrow the search a little, and exclude some of the “pollution”. Thanks again!
You can type a concrete cell type in the “by” field and you can (sometimes) get these cells (e.g. Dm11). It will definitely narrow the search reults
@Krzysztof_Kruk Yeah…if I search “cell identification” by “L1”, for example, then I get results like “pSP5b, put_PDL15f”…which is definitely not the cell type I was looking for.
So…I was hoping for some way to say “NOT pSP5b, put_PDL15f” in the query.
Actually, wait…let me just go try that, lol. ![]()
Unfortunately, this doesn’t seem to be working.
However, you can do your search, then export the results as a CSV file with the button near the top.
Then you can open such file in Excel or other Spreadsheet reader and then do some filtering. I don’t know, how to do such filtering, but I’ve sorted the table by “Cell Identification” column and scrolled down, to find the right results:
Then you can copy the corresponding root Id and open it in FW.
Edit: Heck, you an even copy all results (such file will have 8.9MB in size) and then do any searching in Excel
@Krzysztof_Kruk That…is brilliant! Thank you, sir! ![]()
Lol! I just downloaded the entire database as a CSV, lol! I’m gonna run my own queries now… ![]()
Thanks, KK.
Yes, have already used that database finding T3 cell and will use it more for sure trying to find other examples
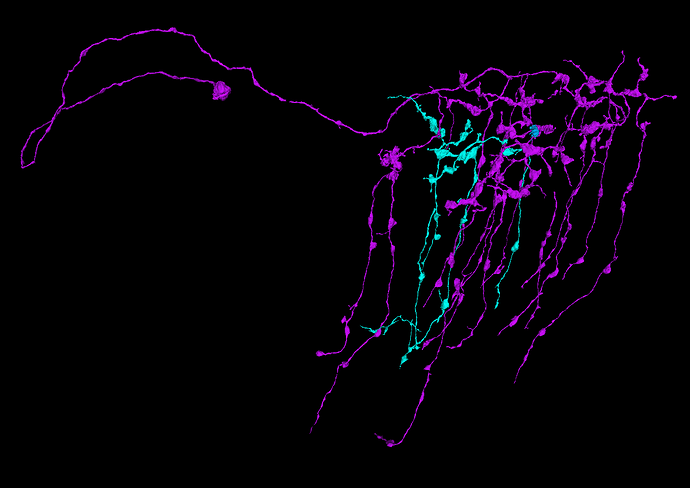
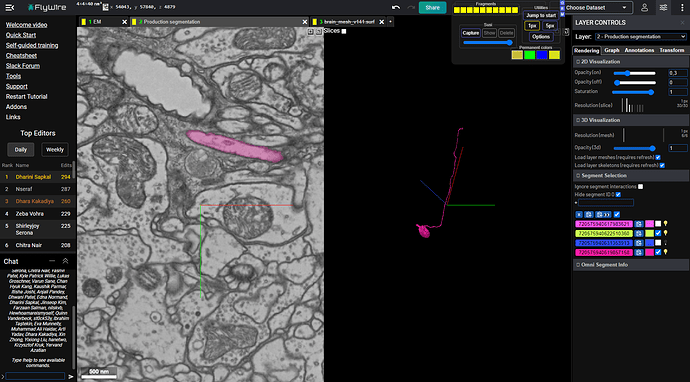
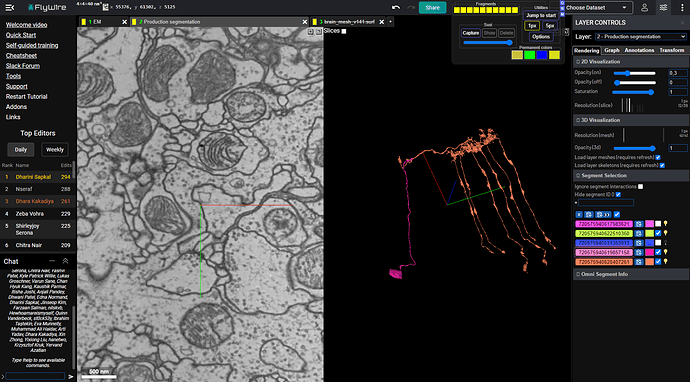
I’ve taken one of these incomplete cells (teal) and found a small misaligned fragment (blue) and then found another misaligned fragment (purple) relative to that small piece.
Here is the result:
https://ngl.flywire.ai/?local_id=6a737c6bd800184afc3b90da1a714501
So looks, like it was worth a try ![]()
I’ve found a new way to find some cells and informations about the dataset.
Go to: https://catmaid.virtualflybrain.org/ and click on the “Adult Brain (FAFB)” link. You’ll be taken to the CATMAID tool.
Click the “Adult brain” image and you should be taken to to a viewer with out dataset (FAFB) loaded:
You can browse it the way, we do it in the FlyWire, by scrolling, ctrl+scroll, etc.
Then you can click on the “Tracing tool” icon:
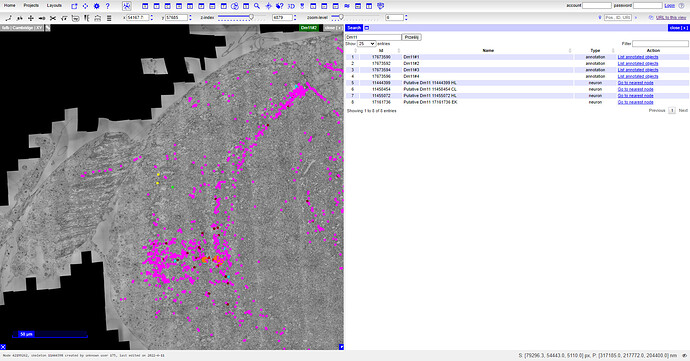
There will be a lot of tools. Among them, there’s a “Global Search” tool:
There you can find all kind of information. Unofortunately, I wasn’t able to find any of the ones missing in our spreadsheet, but I’m sure, that information is different, than ours.
I don’t fully understand the interface (I see, there’s a 3D view for example), but what understood is, that when you found a cell and it has “Go to nearest node” in the “Action” column, you can click on that link, and will be taken to the correct coordinations:
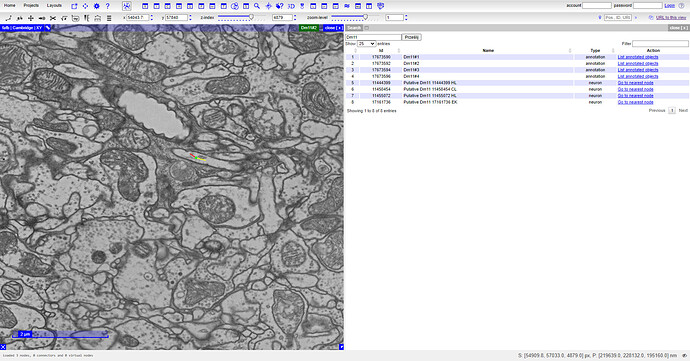
I’ve clicked on the first “Go to nearest node” link (5th row) and was taken to the view visible on the left. You can now zoom in to see, where exactly is the green point. After that, check the coords at the bar above (x, y, z-index) and you can copy them one-by-one to FlyWire. You’ll be taken to the same place.
Zoomed in:
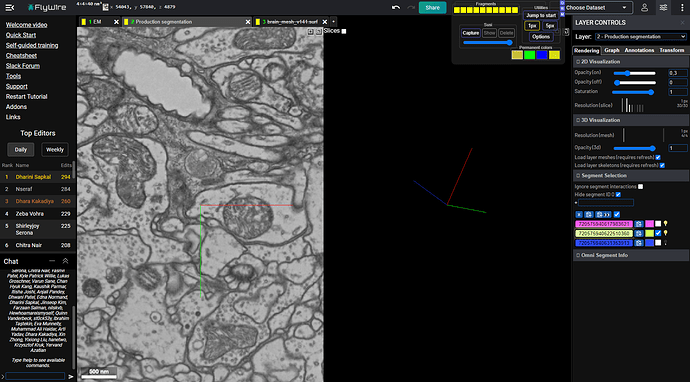
Same coords in FW:
And with the same fragment selected:
And when we add the missing trace, it’s indeed (probably) a Dm11 cell:
None of these fragments are identified in our database, which proofs, that CATMAID indeed has different data ![]()
You can also copy CATMAID IDs and paste them in the search bar in the top right corner (to the left of “URL to this view”. It sometimes takes you to the correct location.
Also, when you browse our database, you can often find entries like “Catmaid ID: nnnnnnnn”. That number, when searched in CATMAID will take you to the correct position.
@Nseraf There’s also a plugin for Blender, which allows importing neurons directly from the CATMAID. I was able to import some segments, but had problems with others. Maybe you’ll have more luck.