I just tested your replacement link and can ID just fine with it! Glad the problem’s been found and fixed. ![]()
Finding downstream partners of the Horizontal System and probably also Vertical System neurons seems to be a good way to find many T cells.
After opening all the parners of one of the HS cells and removing most of the other types, I found over 500 of T cells. Many of them still unfinished. For T4 and T5, all of them are probably of “a” subtype and for VS cells all will probably be “d” subtypes. Didn’t find a way to search for the other two types.
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5933764844716032
Some might be cells of other type. I didn’t comb through them.
When identyfing at random like it looks like you do, how do you know what cells have already been classified or can you risk doing the same job several times?
And are you classifing cells that are also unfinished?
Currently I only identify the cells I completed (although, some of them could also be completed by other players). And even then, I almost always check the status of a cell before submitting its identification.
I classify only the cells, that are completed or, at least, marked as completed (lighbulb yellow, but I already marked them as complete).
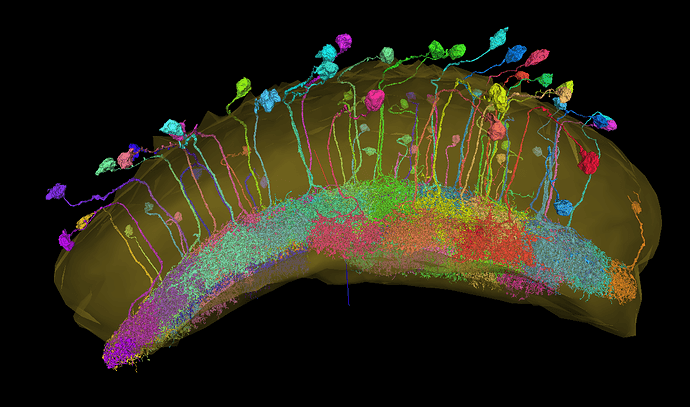
Be careful, when using the neuropils’ 3D models for recognizing layers in the medulla (at least the left side).
Here’s a screenshot of a bunch of Pm4 cells. They all arborize in M8 and M9 layers, but, as you can see, the neuropil model matches it only in the right side:
The groove around Medulla is M7 also known as the serpentine layer.
On the right side the Pm7s are at the correct spot, but on the left side, they are even below the 10th layer of the model.
Source of the Pm7s: FlyWire
To find all, or at least most of the cells of the same type, one can do the following:
- Select a few (lets say, 4-5) completed cells of the given type, preferably from different places in the brain.
- Using the Connectivity app collect all the upstream and downstream synaptic partners’ IDs of the selected cells.
- Go to a site like molbiotools.com, where you can add many lists and find all the common elements between them.
- From the result, get the cell(s) ID, that exists in all of the compared lists.
- Again, using the Connectivity app select all the downstream and upstream synaptic partners of the selected cell(s).
- You’ll probably get thousands of IDs, but they should contain the cells you were looking for.
- Paste the resulting IDs in the FW, preferably in batches and find the correct cells.