Thank you! I’d be more than happy to assist with drafting too, or creating more breakdowns. I’ve just been slacking on them lately ![]()
No problem for me either. It would be cool to have this knowledge condensed in one blog post.
Thanks all! I will wait till month’s end to get started, so that Nik has time to come back from vacation and TR77 has time to reply here. This will be a cool little project to start November though.
those are wild looking!
A little bit of info about all those Lcn, Lccn, Mt, etc. cells. Those cells are known as VPN (Visual Projection Neurons).
We have 2 general types here: columnar and tangential.
Tangential types:
- Mt (Medulla tangential)
- Lt (Lobula tangential)
- LPt (Lobula Plate tangential)
Columnar types:
- MC (Medulla columnar) (not shown in Fischbach)
- LC (Lobula columnar)
- LPC (Lobula Plate columnar)
- LPLC (Lobula Plate-Lobula columnar)
- LLPC (Lobula-Lobula Plate columnar)
The last two are also known as Lccn (Lobula complex columnar neuron).
In the Fischbach’s paper we have 2 Lccn cells:
Lccn1 - its other name is LPL or LPLC
Lccn2 - also known as LLP or LLPC
Because at the time, the Fischbach’s paper was created, there was known only one type of the LPLC neuron, it was grouped with the only one known LLPC type, hence the Lccn group with its two types. Currently, there are known more types of both Lccn1 (LPLC) and Lccn2 (LLPC), and these names aren’t usually used. Now we are using either LPL or LPLC and LLP or LLPC names.
In the same paper there are also Lcn cells, which are just other names for LC neurons (also known as LCN).
Here are subtypes available in each category:
Mt: Mt1 - Mt15, Mt-VPN1, ChaMtnew1, vGATMtnew, vGlutMtnew1, vGlutMtnew2, Mti1, Mti2, Mti-DRA1, Mti-DRA2, OA-AL2i3
Lt: Lt1 - Lt12, Lt21 - Lt33, Lt51, vGATLtnew1, vGATLtnew2
LPt: FD1, FD3, FD4, H1, H2, H5, HS
MC: MC61 (LC10C), MC62, MC63 (VPN-MB1), MC64, VPN-MB2
LC: LC1 - LC27, LC10A, LC10B, LC14b, LC33, ChaLcnnew1, vGlutLcnnew2
LPC: LPC1
LPLC: LPLC1 - LPLC4, LPL01 (type 1 LPL) - yes, there are both LPL(C)1 and LPL01
LLPC: LLPC1
There are probably other subtypes of these cells. These are the ones, I’ve found.
There are also other types of VPNs, like: PDH, S1, V1, V2 and many others (see the tree here: http://flybase.org/cgi-bin/cvreport.pl?cvterm=FBbt:00048286&childdepth=2)
Basing on the knowledge, I’ve recently gained about the giant neurons in the Lobula Plate, I’ve created a sort of a “tool” to help recognizing subtypes of T4 and T5 cells.
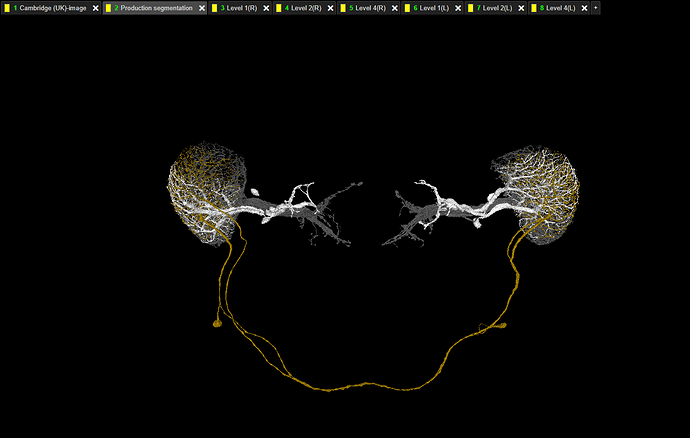
It’s in this link: FlyWire (link has been updated and now contains all six Horizontal System neurons - it’s the same one, as a few posts down below).
After clicking on it, you should see something like this:
At the tab bar you can see additional tabs - from “Layer 1(R)” to “Layer 4(L)”. Each layer represents one layer in one of the Lobula Plates.
To use it, just add your T4 or T5 cells in the standard “Production segmentation” tab. Then hide all the layers from the Lobula Plate opposite to the one, you’re be working with (e.g. if your cell is in the left Optic Lobe, hide all labels with (R) in the name).
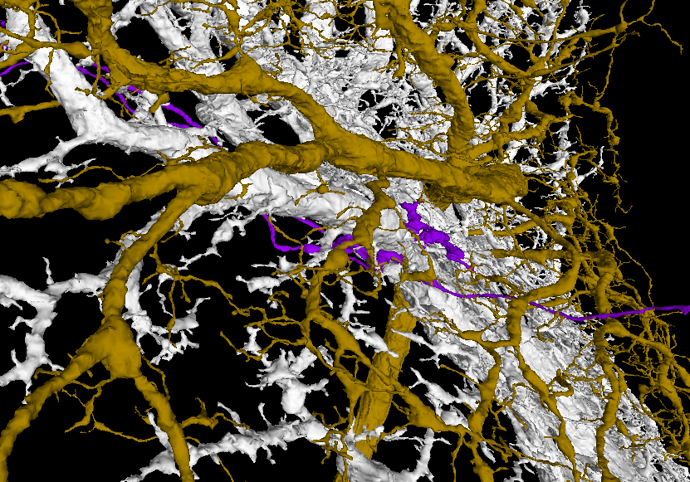
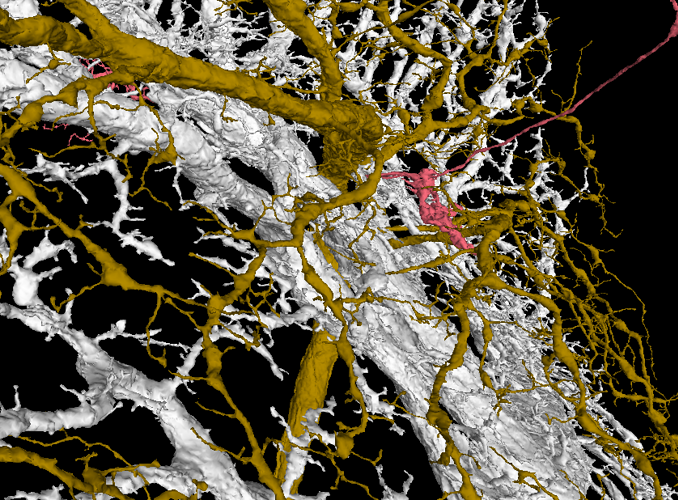
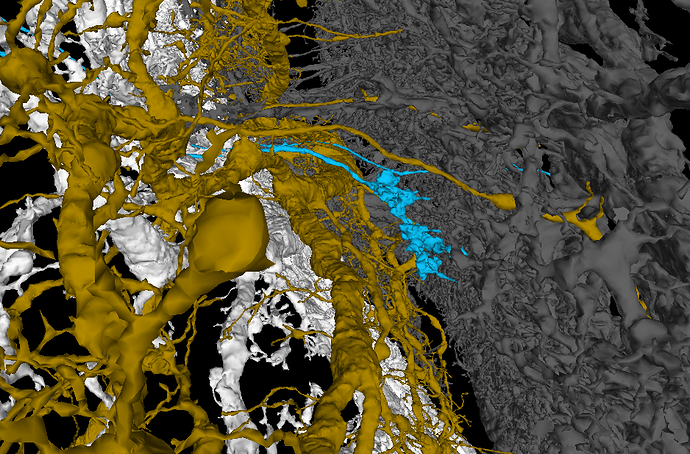
The white layer are Horizontal System neurons and they arborize only in Layer 1.
The yellow-brown layer is a H1 cell - Layer 2.
Layer 3 is empty.
The gray layer are Vertical System and Vertical System-like neurons - Layer 4.
The easiest way to work with the layers for me is to add a T cell, hide corresponding Layer 4 and check, where the axonal part of the T cell is. You can easily tell, if it’s in the Layer 1 or Layer 2. If it float above the two layers, turn on Layer 4 and determine, if the axonal terminal is in that layer or in the empty space between (Layer 3). It works suprisingly well. I had only problems with cells near the borders of the LOP.
Here are some examples of each subtype for T4 cells:
T4a
T4b
T4c
T4d
Instead of Horizontal System neurons you can use Horizontal Centrifugal cells to have better coverage of the Layer 1 (white), but they are much denser and are “heavier” for your computer.
Also, be careful with the H1 cells (yellow-brown), because they have arbors in both Optic Lobes, but only the arbor nearer the soma arborizes in the Layer 2, so be sure, that you’ve turned off the other H1 before classifing your cell.
I’ve chosen the colors, I’ve chosen, because I wanted them to be different from anything a cell could get and to have each layer diffrent color from the others.
Oh, this should be great for identifying batches of T4/T5s for me! I’ve been putative ID’ing as main types as I’ve been a bit more focused on getting more done than worrying about spending 5 minutes IDing each one, but this should make it loads easier to get more accurate IDs.
Indeed. In the last couple of minutes I was able to identify 20 T4s, most without much doubt. I’m surprised, the layering is so visible.
Very interesting way to to it, i am wondering if there might be a possibility to do something like this for other cells also, like using some of the Dm cells too help identify Tm/TmY/Mi cells
will link to this post from the “different cell structure tab” and add some of the info directly into the sheet
I believe, there should be a way to do it both for Medulla and Lobula. There are wide-field cells in both of these neuropils.
So the one problem I am having with this is technical - I always get an error trying to submit an ID on a neuron (“Coordinates outside of the segmentation.”) when I’ve got the coordinates pointed properly in the neuron. Not sure why it does this, so I guess I’m going to have to submit the IDs through a different tab or manually via the menu.
It’s still GREAT though, super helpful.
I have this problem since yesterday. Not sure, if it’s related in any way to the Giant Neurons.
Edit: I guess, the new version of the segmentation, that @M_Sorek mentioned, might be the reason for the problems with submission.
Anyway, when I’m deciding, which neurons are which, I usually take a whole bunch of them, then use the Classifier addon to separate them and then label them one by one. I copy appropriate identification and then just: - find correct point, - select the Indentification dialog, - paste the label, -OK. So even, if the Neurons are the problem, you could do the same, then remove all the tabs and identify them normally.
i have had this problem once in a while, but not in the last few days, no idea what the reason is, but even when i have used the same point both to complete and submit identification for a cell it can happen. usually it work if i move the point a fraction and try again. since it often happen several times on the days it happen and then a long time too next time. it could be something as simple as the server beeing slower to respond for some reason when it is happening.
Link updated with the last two HS cells newly found by @AzureJay and me:
FlyWire
@annkri
I had some error messages before, but this one (about coords outside the segmentation) was a first for me.
Sorry to hear that folks are encountering an issue with the crosshairs for labeling/identification. As annkri mentioned, I’ve had it sporadically occur too as well. I agree that it might be something with a slow server response since usually repositioning the crosshairs fixes it. However, in order to help us track down the bug, if folks could send us a bug report saying it’s through the “Submit Cell Identification” menu or through KK’s addons, etc. that’d be very helpful!
@KK Just fyi v15 has no affect on segmentation on FlyWire itself. It is a separate resource that was recently suggested to us to share with folks as a possible aid for misalignments especially since there are a few bad cracks/jumps in the right optic lobe. (For example, you’ll notice that the v15 link is missing some of the usual FlyWire options such as Sandbox/Production and lightbulb buttons due to v15 being its own separate dataset in neuroglancer)
Wow this is a clever design! Awesome resource.
I’ve tried to submit an identification, but had no luck.
I’ve tested at multiple points inside a cell and multiple resolutions.
All addons turned off.
The submit url was: https://prod.flywire-daf.com/neurons/api/v1/submit_cell_identification?valid_id=720575940611509966&submit=1&location=43204.60546875,19660.46875,5478.9169921875&tag=T4a%3B%20FBbt_00003732
Selected location: 43204, 19660, 5478
RootID: 720575940611509966
Identification label: “T4a; FBbt_00003732”
Error message: “Coordinates outside of the segmentation: [[43204 19660 5478]]. This might be a scaling issue.”
Happens to any other cells.
Refreshed at least a few times for each try.
Not even one success submit since at least Saturday.
Can’t complete cells too.
Thank you, KK! I’m passing this report along to the devs. I’m wondering if something is up with the lightbulb menu since you are unable to submit completions as well…
Nope, the menu displays correctly. After clicking appropriate options, correct dialogs are shown and after submitting, I’m redirected to the correct pages. The URLs of those pages seem to be ok too. Also, no errors in the console on both pages (FlyWire and submission result page).
Problem has been fixed (thanks @M_Sorek for solving the issue!). There was something wrong with my settings/state. Because of that, also the link above (with giant neurons) have been “poisoned” with the bad settings. I’ve recreated it with the correct settings and replaced the old one. Now everything should be ok ![]()