Yes, you’re right. I didn’t take that into account, that there might be a difference in the number of synapses per cell.
I’ve also read (only now), that authors of the paper analyze the synapses only for a single branch of the cell with 600-1000 input synapses and 100-170 outputs. This is probably the reason, why the cells on fig. 1 and fig. 4 look different and it’s also possible, that this is the reason, why they didn’t have many T4b/T5b synapses amongs their inputs. There’s also a possiblity, that our main cell have some mergers or that I’ve miss-labeled some T cells.
All in all, I’m more convinced, that this might actually be the LPi1-2 cell.
What do you think?
i agree with it beeing LPi 1-2
and as it is written under picture 4 it incomplete due to the study beeing performed only on a smal area of the optic lobe. Only LPi 3-4 is complete in that picture.
Great, thanks for all the help. I’ll label it then as LPi1-2 and move on to Y cells.
Actually, I was going to do the TmY cells, but when I’ve started with them, there were many other cells between the ones I wanted, so I’ve moved to them, to eliminate them from further searchings and that’s how I’ve ended with labeling LPi and Y and probably Tlp soon. When I finish them, I hope, I’ll be able to go back to the TmY again, then Tm as my target.
On that case maybe it might be useful to make some threeshold value on the list of synaptic inputs. i have no idea what that threeshold should be, but my understanding is that any cell that have only a few synaptic connections often are mergers.
good luck that is probably some of the most challenging types to label, and my guess is that we will find that many of the different types are actually the same. Or have only minisculle differances like who they connect too.
I could do the threshold, however, there might be a issue with small cells - they only have a few synapses at all. I would have to check it before making such modification.
And I agree, that Tm cells are the most difficult. I’m already having trouble with TmY cells, where many are very similar or have characteristics of a few types at the same time (e.g. medulla part looks like TmY1, lobula part like TmY2 and LOP part like TmY3). And there are probably some types not yet identified. That’s why I’m leaving them to the end to have the best understading of general cells morphology and possible differences within types and to narrow down the search results.
I should add, that there will still be quite a few types not yet identified: for example, many in lamina (thanks to everyone who’s taking care of it!, it’s my least favorite part of the brain) and Li (another difficult group), Mt, Lt, Tl and, of course, many in the central brain. So, still a lot of work ahead of us.
and very little in the left lope is identified so far.
We also have a lot of AM/ Lai cells in both lobes that is very easy to identify but very difficult to complete left, since they are mostly in smal bites a lot of them are not completed or missing big chuncks.
Right, I completely forgot about the other lope, lol.
The other type, that’s easy to identify but hard to complete are Dm9. I really hope, the other researchers will do them some day ![]()
Oh, there’s also one cool type in lamina called Lat. I wonder, how they look like as a whole group.
not sure if i remember how Lat looks like unless it is a case of same cell having different names?
It’s not in the Fischbach paper, but you can see it here: Visual Cell Type Illustrations on the first pic (near the right side) and I’ve also seen them a few times during my explorations.
Edit: sorry, it actually exists in the Fischbach’s paper, I didn’t notice it at first:
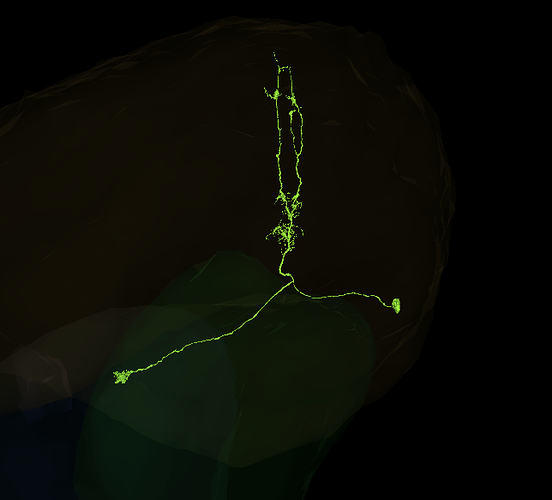
Here are some labeled Lat (lamina tangential) cells! ![]()
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5822216144945152
Awesome!
I didn’t realize, they span all the way to the CB. Also, I thought, each of the main neurites between M and La will go separate way, not as a single bunch.
I have another “type doesn’t exist” problem.
I was able to identify Y1, Y4, Y11 and Y12 cells.
Y2, Y7-Y10 don’t exist.
That leaves us with Y3, Y5 and Y6.
I’ve found almost 300 Y cells other than those already identified, but they all look the same:
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5086231706730496
According to the Fischbach’s paper they should be most distinctive in their lobula extensions, but I don’t see any differences.
Also, of these 3 types, Y5 and Y6 are mentioned only in the Fischbach’s paper.
So, I’m starting to think, all these three types are a one type.
i am not able to see any differance either, but it do look weird that it it 300 cells left in one type when the other Y- cells have only 60-90 cells
I agree, that looks a little unusual, however, I’ve seen similar behaviour elsewhere. For example, some Mi cells are one per ommatidium, so about 800 cells, but others are only one per two columns or even one per 10 columns.
What suggests me, that it might be a single type, is how they look both in the medulla and in the LOP - they fill the planes nicely, without overlapping or creating patterns constructed of 2-3 cells.
I’ll look at them some more, maybe I’ll find some more info to help me decide, what to do.
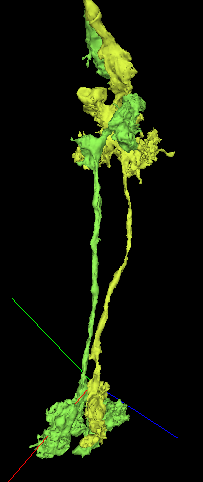
While going through my T-cell farms, I came across this previously completed but unidentified neuron. It has most characteristics of a T2 or T3 cell - I’d be more inclined to call it a putative T2 because it stretches further than a T3 would - but there’s clearly something different going on with a split here in the medulla and a lack of expected branches. Any thoughts? I’ve tried looking on the forums best I can but “T2” is too short a search term and we’ve posted so much over the past year!
Edited to add another example cell to the link below -
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5715177741746176
It so happens, that I’m currently working on T2 cells. If we assume, that everything, that has big-small-big arborizations in the medulla are T2 and every other type is a T2a, I’d say, yours is a T2a.
Here are all T2/T2a cells, I’ve found so far (probably some others between them, I still have to filter them out): FlyWire (warning over 1000 cells, almost 30k chunks). You can see, there are many with this split top branch.
Thank you! So it seems like I’m on the right trend with T2-type. I feel like there might be more distinctions here than T2 and T2a that just haven’t been investigated yet. That of course will take more time and research to figure out, if so, but I will label these putative T2 types ![]()
Im having a bit of trouble deciding between two cells to label as an L1. Tbh im not sure if maybe both of them are, but there should only be one per column, so ![]()
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5365430300442624
I’ve added to the link the L2-5 for the column and a “proper” L1 from another column for reference, with annotations to clarify.
If I could take the lower cluster from one and the middle cluster from the other, i’d be happy ![]() Maybe this column just has two malformed L1s?
Maybe this column just has two malformed L1s?
(Edit: Found a second pair, only these two pairs across the whole lobe though. FlyWire)
I’m going to ask around, but it could be that these are just some odd ones. I couldn’t find any extensions that would indicate that one of these was a different type of cell (ie. centrifugal cell, etc.).
We know that for the photoreceptors (R-cells) that “because of neural superposition during eye development, there is atypical number of photoreceptors at the equator and edge”. While I’d say the second pair you shared is closer to the edge of the medulla compared to the first pair, this could explain why there may be an extra L1…