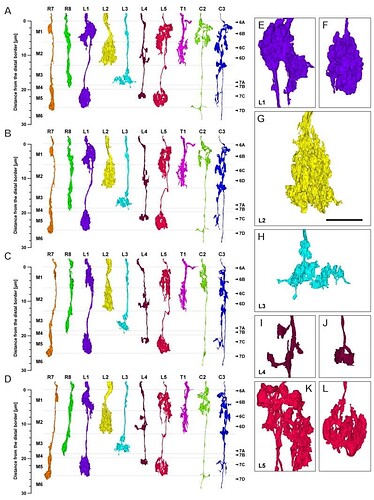
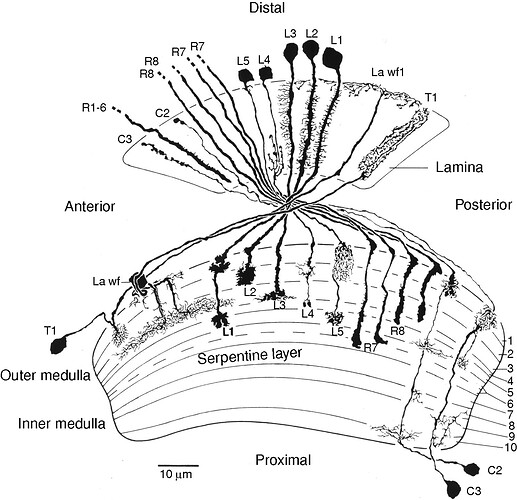
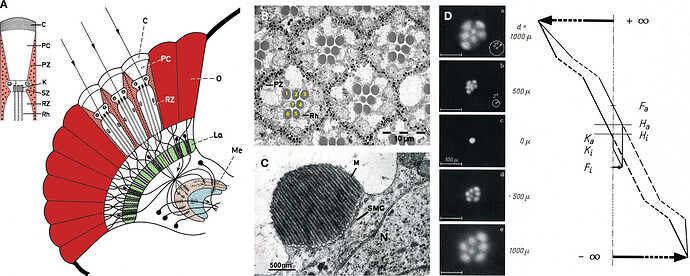
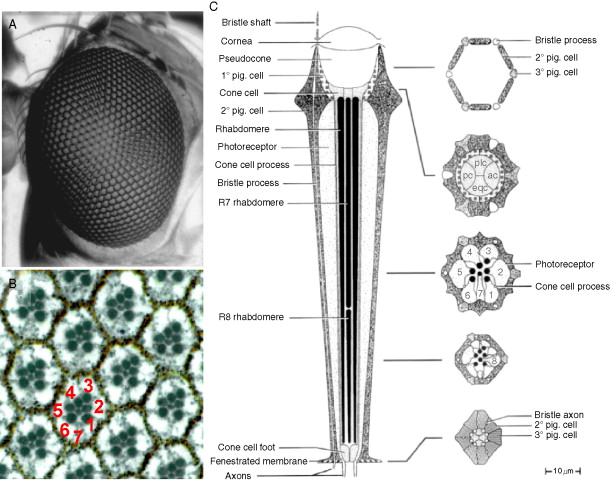
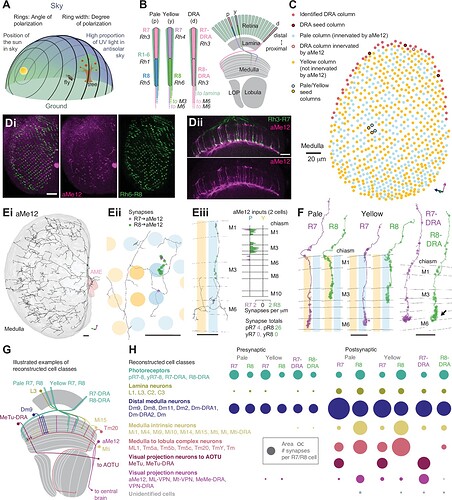
Going from illustrations in:
(Thank you @AzureJay ) and https://www.pnas.org/doi/10.1073/pnas.2025530118
and
(source: https://www.cambridge.org/core/books/abs/behavioral-genetics-of-the-fly-drosophila-melanogaster/anatomical-organization-of-the-compound-eyes-visual-system/8BD880CF0E0AA68EF0782E238C9682D5)
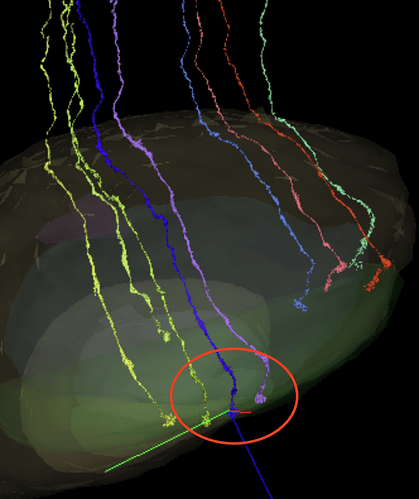
I’m making a hypothesis that the shorter the length of the axon the smaller the number (R1-8), so I’m thinking:
R1
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4576471385374720
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6366956009553920
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6335490299002880
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4774040048762880
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5683473268342784
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6042194339168256
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4718810787479552
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5698150840926208
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4932346092453888
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4627705001672704
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5353413789351936
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5707995577057280
R2
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5773884804562944
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4779279741091840
R3
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6603627397382144
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5658768952000512
R4
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4890495369084928
R5
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4959581126000640
R6
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5731199280480256
R8
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5973594072940544
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6332962576531456
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5407152252387328
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5982268296265728
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6540437120614400
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6481381748113408
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5681822717444096
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6536544026361856
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6749139379421184
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5844710694322176
R1 or 2?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6217767770390528
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4643243253825536
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5618275832561664
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5769143160668160
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6750594727411712
R5 or 6?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5624694820569088
R2?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5898259407044608
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6085481032843264
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4653096177238016
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6561627683946496
R3?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6744175739404288
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5356783619014656
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4959399260979200
R4?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5226430531108864
R5?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5139421338796032
R7?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5572314850656256
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5418628572774400
R1-3s?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4511989053259776
R6-8s?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6544528479617024
Lines 25-71 in gsheet.