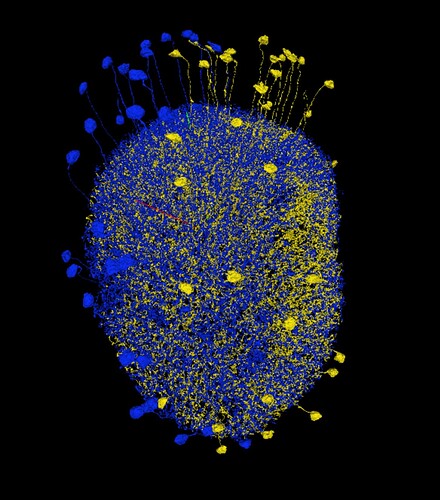
Thank you! I didn’t find that particular info about dendrites density.
As for the layers, also other types of cells tend to have their terminals in these layers, e.g. LPLC so I was hoping, that it will be visible in 2D, but it isn’t unfortunately.
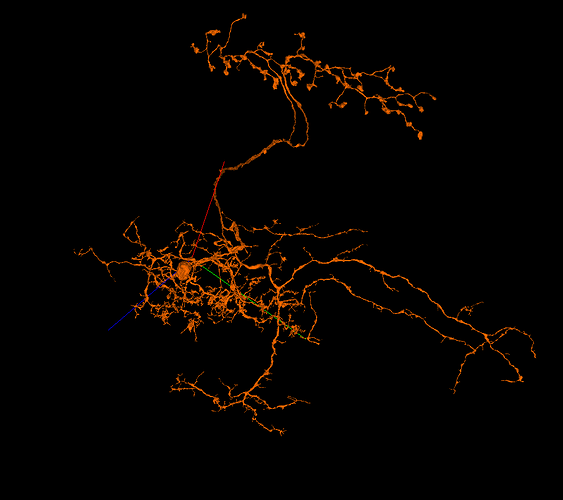
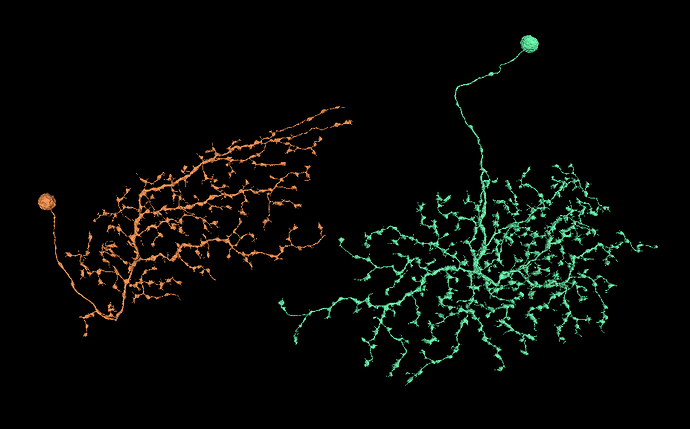
I’m looking for some wide-field cells in the Medulla to help with identyfing layers, like in LOP. While doing it, I’ve read about ML-VPN1 cells (among others). As a side quest, I’ve decided to find all or at least most of them and identify them. And here’s my question: do you think, the cells, I’ve found, are ML-VPN1 cells?
My cells: FlyWire
(the red neuropil is posterior lateral protocerebrum (PLP))
Sources for ML-VPN1:
http://flybase.org/cgi-bin/cvreport.pl?id=FBbt%3A00052138
I would concur that these are likely to be ML_VPN1s. I might check to make sure they’re synapsing with R7/R8 cells as that seems to be a key quality of them, but structurally I’d agree with you!
Thanks!
I was 80% sure, they are the correct ones, but needed a confirmation.
And the do synapse with some R cells, indeed. Additionally, when I’ve checked them, it looks, like they upstream synapse also with Mi4 and Tm9 cells (among others), so it might be another way to tell those Mis and Tms apart from others in their types’ groups. Example:
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5930285585661952
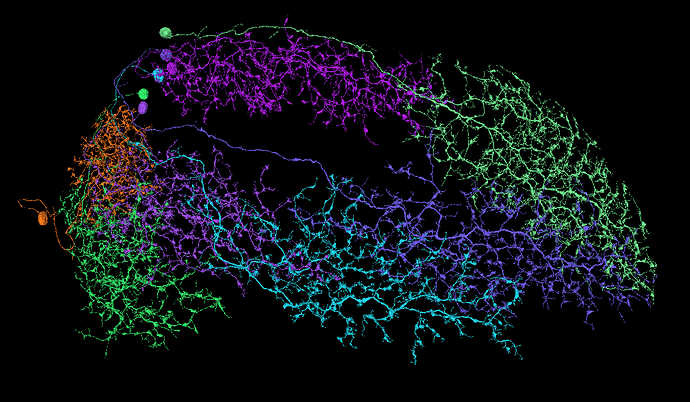
Now, to find all the rest of the MLs. There should be about 65 of them per optic lobe. So far, I’ve found only 26 and had no luck findind the rest:
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5926983628226560
A side quest during a side quest xD
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5581160813953024
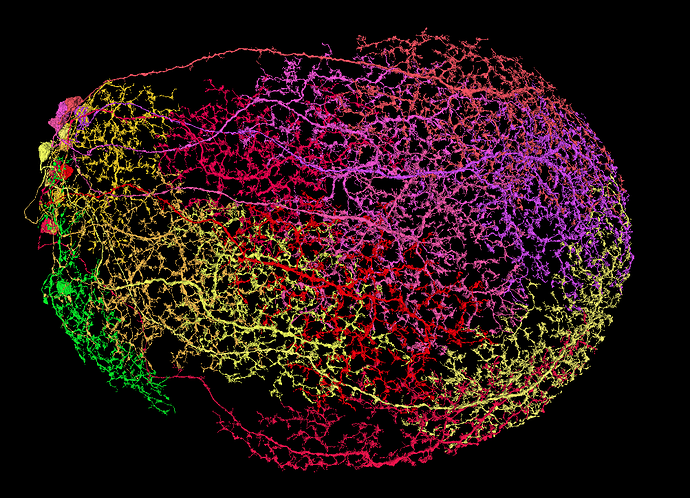
Unfortunately, I wasn’t able to find, what type of cells those are, so I’m leaving it here. I’ve asked ChatGPT, be he also didn’t know, lol. Maybe some of you knows.
i am pretty sure that is a Pm, they are some of the few running a lot of parallel branches into a flower. do not know the original source but according this discussion. we have earlier found out they had too be Pm2 probably because the flowers go down in medulla instead of up like pm 1 and 3
but according to
Varija Raghu, Shamprasad & Borst, Alexander. (2011). Candidate Glutamatergic Neurons in the Visual System of Drosophila. PloS one. 6. e19472. 10.1371/journal.pone.0019472.
pm 2 is supposed too have arbours only in medulla layer 8 and 9
Yes! You,re right. I’ve (wrongly) assumed, that all the Pms and Dms have all their terminals only in the Medulla, so the Lobula part fooled me.
As for the arbours - the more I read the papers, the more I see, that many of the informations are based on previous articles, all the way back to the Fischbach and even before. And the technics used aren’t always very reliable to show whole cells. As far, as I can tell, the research we’re doing in FlyWire is one of the first ones showing not only microscopical or skeletal images, but full 3D look, so there might be some mistakes in the older sources.
Even in the Fischbach’s paper the Pm2 is the only Pm/Dm cell, that doesn’t have it’s soma shown, suggesting, there’s something more there.
I’ll mark then as Pm2s then. Thanks ![]()
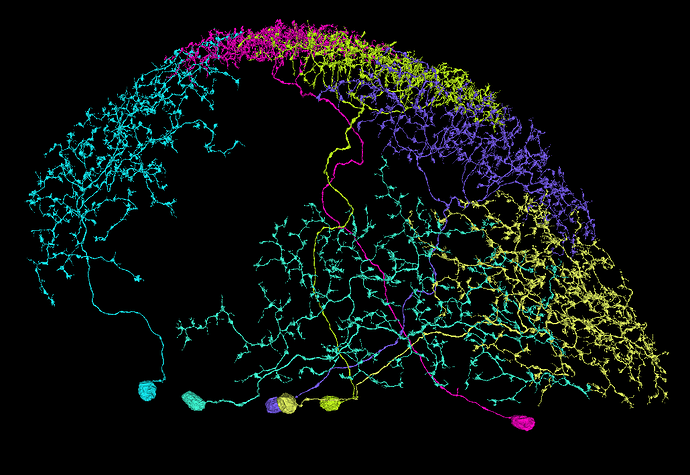
I have trouble classifying this Lawf cell. It has characteristics of both type 1 and 2.
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5839491856072704
I thought about it too. I believe, they are joined on the biological level (as opposed to the data-preparing level), because the “top” two branches are apart like this. If they were separate cells (and only joined during the preparation), the axonal part would be probably intertwined.
found a second cellbody on it
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6186604267831296
Oh, nice, thanks! ![]()
Edit, so it looks, like we have a pair of conjoined neuronal twins xD
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5934680511610880
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5619219525795840
What do you think:
- Are those cells in both links of the same type?
- What is this/are these type(s)? (my guess is Pm2)
I’ve differentiated them that way, that the cells in the second link have at least part of the their soma neurite go “under” or proximal to the main layer of the synapses, while in the first link the neurites are wholy “over” that layer. I did the differentiation, because I thought, over 100 of the same type of a wide-field cells is a bit too much, but it’s just a thought, not based on anything.
One could also try to find differences in the length of the “buds”. There could be 2-3 different lengths, but it might depend on the position of a cell inside the medulla.
i agree with them probably belonging too two different layers since they are making complete layers (might be some that is classified wrong since it looks like the other layer is thicker where you have missing cells in the other.
i would say some kind of DM cells but have not studied who of them it could be, but that also make the chance bigger on it beeing two different layers since we have a bunch that is difficult to find pictures of
Pm have lots of parallell brances so definitvly not that
Those are in the proximal part of the medulla, so they can’t be Dms. As for the parallelism - yes, at least for the Pm4, that’s true, not sure about other types. I will look for some papers about this topic.
I also have these other four types, which I will be looking for more of those same types and then trying to identify them (all in the proximal part of the medulla):
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5318174262165504
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5036699285454848
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6601583236218880
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6284412081668096
As for the pm we have identified 1,2 and 4 in the spreadsheet all with paralell branches.
If you are sure about this not beeing Dm´s i guess the only other choice is some type of Mt (can´t say any of the pictures looks very close.) Also Mi 11 have CB on the right place and might maybe be a better choice. I am guessing there also could be other Mi cells going in from the side.
As for the other pictures i am thinking in the direction of Mt on them but have not worked on those so do not know them very well.
the red one at the bottom on picture 3 looks like a Mt 4 or 8 but at least 3 different cell types on this picture (red, orange close to the green, and the rest)
Yep, you’re totally right. I don’t know, what I was thinking, saying, those are Pm2s. Especially, that I’ve identified a bunch of them not so long ago. I will look at the Mts.
As for the pictures in the previous post, I was able to find probably all the rest of the cells from the first picture and identified them as Mt5.
Now I’m trying to separate Pm1s and Pm3s, because, if I’m not mistaken, they are looking very similar with the main difference being, that Pm1 have clusers together, while Pm3 have them more separated from each other. But I might be wrong with this. Also, have to find a way to tell apart Pm1 from Pm1a.
Here are my results so far:
Pm1 (probably):
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5625519034859520
Pm3 (probably):
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6325998706491392
Might be a good way to say they are different, but have you found anything about a difference in who they connect too? saying they are tighter or more open might perhaps have some difficult cases?
have you noticed the cb are placed in different part of the medula on your suggestion?
Not related but i find that naming a annotation layer with the cell name i am working on help me knowing where i am when having multiple different tabs open.
Nice find! I didn’t notice that. I mean, I thought, that the soma neurites for one type are shorter, than for the other, but I didn’t notice, that that was just because they were further from my point of view.
I know, that they are all connected to some of the large whole-field cells, but that doesn’t help much. Checking for the smaller synaptic partners is a great idea.
As for naming, I’m using a Chrome addon called “Rename Tab Title”. It allows you to rename a whole browser type, no matter, what’s inside that tab. It stays even after refreshing a tab or closing the browser. To rename a tab, one has to simply press Ctrl+Shift+E and type the new title. Helps tremendously, especially, when trying to split a large group of similar cells between types and moving cells between the tabs.
Following your advice, @annkri, I’ve started to check the synaptic partners of the cells in the question. At the last moment, I’ve changed the strategy a little bit. I was still using the Connectivity app, but instead of checking the partners, I was looking at the neurotransmitters at the synapses. I found out, both types of cells were about 90% on average GABAnergic on their outputs, but there were noticeable differences in the other neurotransmitters. All the Pm1s have almost 0 adrenergic (Ach) synapses, but Pm3s have always a few percents of Ach (clearly noticeable in the charts).
Using this method, I found between the Pm1 cells one, that didn’t belong to any of the groups. Belong the Pm3s there were two Pm1s and one cell I’m strill strugling with classifying.
So, I’d say, it’s quite a useful method for checking similarities between cells.