Thanks!
To be honest, I’m starting to think, TmY3 and TmY8 are duplicates. It’s a problem similar to Mi8 and Mi9.
So far, I’ve found 314 cells and don’t see any differences significant enough to mark some of them as a separate type. So either they’re the same type or the other type is so good at hiding, that I didn’t find even a single instance of it.
My guess, at least some of the differences in the Fischbach’s paper can be explained by the fact, that both LO and LOP are concave so cells near the edges will look longer, than the ones in the centers.
Reading the definition of the cells i think much like you. i think the only thing making me belive it is two distinct cell types is the way TmY 8 branches in lobula compared too TmY 3 having only a straight branch. Every other description i find looks very much like it is only shifted one layer up or down but have the same basic outlook.
I have also read that some of the neurons can have slightly different look based on age of the sampled specimen (adult/not adult etc) this might maybe be enogh to count for small differences?
Some of the cells in my last link near the edges have their lobula branches squished and they are not straight anymore, so that might be a source of further confusion during light microscoping.
Yes, the neurons in different stages of life of a fly look very different. For example, we’ve seen recently the larva has only 3000 neurons in its whole body. There are also different pupal states. Not sure however, if there are many differences between different adults and both Fischbach and FAFB are done on an adult, afaik.
For now, I’ve marked all the cells as TmY3 and moved on to search for other TmYs (currently TmY4). When I’m done with most of the types, maybe I will be more able to tell, if some of the still unmarked then are TmY8 indeed.
Thanks for your input.
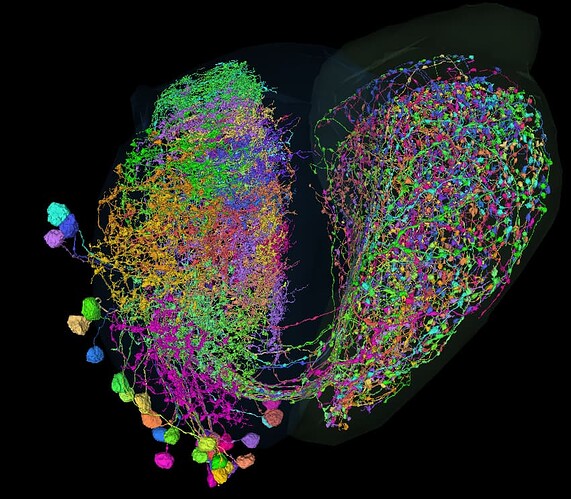
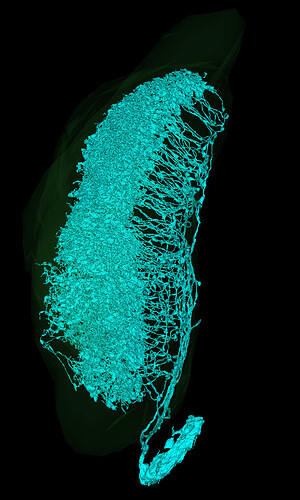
Can any one tell me, what this type of cells is?:
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6181339046871040
The only ones, that go from LO to LOP are Tl cells, if I’m not mistaken, but both types in Fischbach are quite different and FlyBase says, that there are only two types of those.
There is a cell named LLPC i have not been able to find any pictures of this cell so could be that it also go into the central brain, but based of the name it should have its cb in lobula and go into lobula plate
That was my first thought. However, it looks, that we already have these cells in the spreadsheet and they look quite different.
then i am not seeing what else it can be than a unknown Tl cell type
yep. I’m starting to think, there are more unknown types, than known types, especially in lobula and lobula plate. Most, if not all research papers look at the optic lobe from the perspective, that we see in the Fischbach’s paper. However, there are are multitude of cells, that look flat from that perspective, but can be easily differentiated from other views.
Also, as in the case above, if some of the main branches connecting parts of the cell are somewhere outside the main neuropils, the parts of the cells are treaten as separate cells.
I have another problem: I want to find LPi1-2 cell(s). The only source for this type, I found, is here: Neuronal circuits integrating visual motion information in Drosophila | bioRxiv
Figure 1 shows the result of the light microscopy and figure 4 - a drawing + synaptic partners.
Already here I have a problem, because the drawing in fig. 4 looks different than the photo in fig. 1. I’m assuming, they’ve drawn only part of the cell on fig. 4 to make it look more similar to the other types, but I’m not sure, if such assumption is correct.
The most similar cell in the LOP, I’ve found, is this one:
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5040357815353344
There are also 2 other hidden “full-plain” cells in that neuropil. Not sure, if there are any more like these there.
Here are the synaptic inputs, I was able to gather:
Of course, these aren’t all the inputs, only the ones, that have been labeled, but in this case, the T4 and T5 cells seem to be the most important.
Fig. 4 in the paper says, that there should be mainy inputs from T4a and T5a. And it’s consistent with my results, however, there’s quite signigficant portion of inputs from T4b and T5b. I wonder, if we can assume, the results are similar enough to say, that that big cell is indeed an LPi1-2.
I should add, that I’ve searched through out the whole LOP quite thoroughly and didn’t find anything even remotely similar to neither fig.1 or fig.4 other than that big cell above and cells LPi2-1 and LPi34-12, which are already identified (available in the sheet). I’ve found some other (unknown) LPi types, many LPt and Y cells and that’s basically it for this neuropil.
TL;DR: is the cell in FW link an LPi1-2 neuron or not?
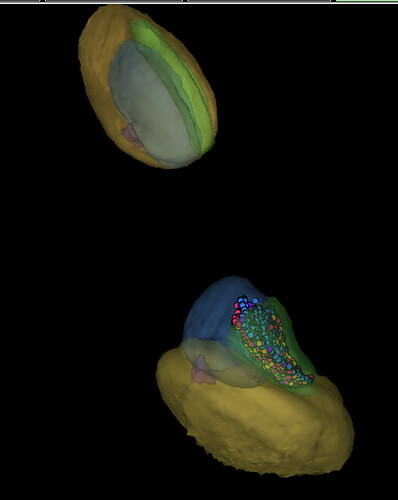
From my understanding in LPi 1-2 incoming synapses red should be mainly in layer 1 from center while outgoing blue should be in layer 2. Like what is seen here. since it is so big i was only able to test this theory on the 8 cells with most ingoing/outgoing synapses.
From what i understand your summary show only how many cells synapses with the cell regardless of how many synapses each cell have, so the 4/5 b could have 1 synapse and still be on the list while each 4/5a have 50 synapses?
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4693636446945280
Yes, you’re right. I didn’t take that into account, that there might be a difference in the number of synapses per cell.
I’ve also read (only now), that authors of the paper analyze the synapses only for a single branch of the cell with 600-1000 input synapses and 100-170 outputs. This is probably the reason, why the cells on fig. 1 and fig. 4 look different and it’s also possible, that this is the reason, why they didn’t have many T4b/T5b synapses amongs their inputs. There’s also a possiblity, that our main cell have some mergers or that I’ve miss-labeled some T cells.
All in all, I’m more convinced, that this might actually be the LPi1-2 cell.
What do you think?
i agree with it beeing LPi 1-2
and as it is written under picture 4 it incomplete due to the study beeing performed only on a smal area of the optic lobe. Only LPi 3-4 is complete in that picture.
Great, thanks for all the help. I’ll label it then as LPi1-2 and move on to Y cells.
Actually, I was going to do the TmY cells, but when I’ve started with them, there were many other cells between the ones I wanted, so I’ve moved to them, to eliminate them from further searchings and that’s how I’ve ended with labeling LPi and Y and probably Tlp soon. When I finish them, I hope, I’ll be able to go back to the TmY again, then Tm as my target.
On that case maybe it might be useful to make some threeshold value on the list of synaptic inputs. i have no idea what that threeshold should be, but my understanding is that any cell that have only a few synaptic connections often are mergers.
good luck that is probably some of the most challenging types to label, and my guess is that we will find that many of the different types are actually the same. Or have only minisculle differances like who they connect too.
I could do the threshold, however, there might be a issue with small cells - they only have a few synapses at all. I would have to check it before making such modification.
And I agree, that Tm cells are the most difficult. I’m already having trouble with TmY cells, where many are very similar or have characteristics of a few types at the same time (e.g. medulla part looks like TmY1, lobula part like TmY2 and LOP part like TmY3). And there are probably some types not yet identified. That’s why I’m leaving them to the end to have the best understading of general cells morphology and possible differences within types and to narrow down the search results.
I should add, that there will still be quite a few types not yet identified: for example, many in lamina (thanks to everyone who’s taking care of it!, it’s my least favorite part of the brain) and Li (another difficult group), Mt, Lt, Tl and, of course, many in the central brain. So, still a lot of work ahead of us.
and very little in the left lope is identified so far.
We also have a lot of AM/ Lai cells in both lobes that is very easy to identify but very difficult to complete left, since they are mostly in smal bites a lot of them are not completed or missing big chuncks.
Right, I completely forgot about the other lope, lol.
The other type, that’s easy to identify but hard to complete are Dm9. I really hope, the other researchers will do them some day ![]()
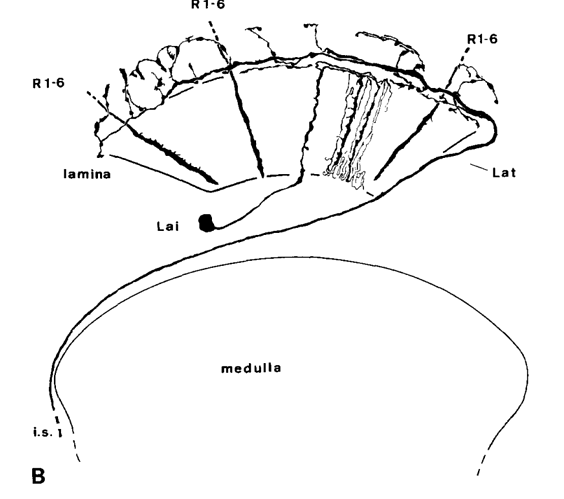
Oh, there’s also one cool type in lamina called Lat. I wonder, how they look like as a whole group.
not sure if i remember how Lat looks like unless it is a case of same cell having different names?
It’s not in the Fischbach paper, but you can see it here: Visual Cell Type Illustrations on the first pic (near the right side) and I’ve also seen them a few times during my explorations.
Edit: sorry, it actually exists in the Fischbach’s paper, I didn’t notice it at first: