That’s a really good idea !
Here my first one :As there’s no “ShowMeMe” action, i use Elypsoid annotations as much as i need to mark all branches i checked.
not sure if it is a rule i bouquet cells, but i have noticed that the one i have been working on had a cluster of branches going in the same direction, and even if i had not found a connection to all of them the first time around, since they followed the direction of the others closely, i followed them up toward the soma again, and found that they belonged to my cell after all.
A tip for dealing with misalignments, thanks to M:
- Try to find a nearby branch of which you have the correct extension through the misalignment. I like to look for big or unique shapes. Looking at organelles may help as well.
- Place an annotation point before the misalignment on that branch.
- Place an annotation point after the misalignment on that same branch.
- You can now click on the annotation points on the right side to jump between them and find the correct extension for your branch.
Looking at the annotation points in 3D also helps to see the direction where your branch should continue. For instance, if the annotation points jump towards the right, and the continuation of your branch jumps left, it’s unlikely to be the correct continuation.
Here’s an example of where I used this technique: FlyWire
Definitely ! I’m in a “bouquet cell” and i find branches splitting every time into new branches. It really can happen everywhere, even if a split has already been found near.
You can slide “Saturation” all the way to the left (to 0) to make all segments grayish white. It helps, if you want to determine, if a given segment’s structure matches the rest of the cell, but the colors are different and make it harder to say.
Of course, you can also change the color of the segment to match the color of the cell, but the saturation change is quicker and changes colors of all the segments at the same time.
"Pinched" Neurites, and the Infamous Row 4 Cell
Wasn’t sure where to post this, or even if it deserves it’s own topic…I’ll let the community decide.
It seems to me that there is a pattern to some of the problems showing up, in the Q&A log. Many of the cells that I have seen there, including the infamous “Row 4 Cell” involve a neurite that suddenly just ends. The pattern I am seeing is that most of them are type L cells, and they tend to suddenly end in the area known as the chiasm (i.e. the “Black Spill” area, discussed in a separate topic), which is essentially the boundary or transition from the medulla to the lamina. As has been documented in the FlyWire blog, and elsewhere, the lamina is essentially a series of modular, repeated structures, known as “Laminar Cartridges” or “Columns”, in which the same cell types repeat the same pattern of structure, over and over (about 800 times, if I’m not mistaken).
Given that these repeated laminar structures show the same basic arrangement of the component cells, each time, it seems that we can make use of that regularity, in predicting the continuation of the dead-end neurites mentioned above. Applying this approach to the infamous “Row 4 Cell” (which shows up again and again, in rows 84, 121, 123, etc.), yields the solution pictured and linked below. In the view below, the row 4 cell is shown in red, next to a nearby cell of the same type (L5), from an adjacent laminar cartridge/column, shown in green. For reference, the L1 cells of both cartridges/columns are shown in blue. As can be seen, the similarity between the two cartridges/columns is sufficient, in my opinion, to suggest a practical solution strategy. This practical similarity can be further reinforced by comparing with several other nearby cartridges/columns, and I encourage others to do so (I’ve spent days now, looking through the dataset and comparing these structures).
Also, it is worth noting that, although these “pinched” neurites do appear to be actually separated/broken/pinched apart, in the 2D EM image data, the data would look the same if they suddenly reduced in diameter to the point that they passed between slices (i.e. not actually broken, just very thin). Either way, it is a problem that seems to come up over and over again, in the same area, regardless of what the actual explanation may be.
If anyone has any thoughts or insight on this, I would appreciate feedback. I have tried applying this approach on several of the cells in the Q&A log, and the results have been very encouraging…enough so that I felt it deserved mention here, and possibly it’s own topic for discussion.
Cheers, all.
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5950851256418304
I had to immediately check out this method last night and had a few successes already! This is great, thank you.
Can you offer any additional insight into finding a neighboring L1 cell in the 2D?
Sure, I was thinking about putting together a guide, outlining some of the tips and tricks I’ve noticed, but figured I’d wait until hearing some feedback from the community first.
For now, you are definitely right to ask about locating L1’s. The L1 and L2 cells kind of form the central core, of each laminar cartridge, and are generally the easiest to spot, once you know what to look for. The laminar cartridges form sort of a hemispherical surface, that appears hovering over the medulla, in our view with the 3D brain mesh turned on. If you look at them in a 2D cross sectional view (i.e. a plane perpendicular to the long axis of the cells), then the backbones of the L1 and L2 cells tend to stand out as two relatively large neurites, touching one another. If you zoom out a little, you will notice the repeated pattern of L1’s and L2’s, spread throughout the lamina (i.e. repeating clumps of two relatively large neurites, touching one another).
In my experience, the backbones are thickest and most recognizable in the dendritic arbor, immediately downstream of the soma. Also, the axonal boutons can be pretty easy to spot as well, once you get used to recognizing them, b/c they are also very bulky and tend to stand out, with the same repeating pattern.
It took me a while of staring at the 2D, before I got a feel for what to look for, and how to recognize them. As I said, I’m thinking about putting together a guide, outlining this approach.
Thanks for the feedback, AJ. Feel free to post or message me, if you have any thoughts.
Cheers.
Yes, please, make a guide. When I’m reading your and @AzureJay’s posts, I feel, like I’m so behind in the knowledge of the structure of the brain. I’m still tracing mostly blindly looking for obvious missing things without knowing, what and where I should look for and how can I help myself by knowing, what I’m looking at.
There are, of course, the scientific papers, but sometimes they are quite difficult to understand and they are often based on datasets other, that our FAFB so it harder to transplant that info to our local environment.
I know exactly what you mean, and how you feel…that’s why I spent the time putting together this approach. With a generalized version of this approach, I feel optimistic that we should be able to address most or all of the issues we have run into.
Thanks for the feedback, KK. Let me know if you have any further thoughts.
Cheers.
As a prelude to a guide, the link below contains several completed and identified L1 cells, enough to give a basic impression of the arrangement of the lamina (each cell being the center of a laminar cartridge). Just imagine several hundred more, filling in the gaps, and you will have the right picture in mind.
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6691690635067392
The first image below, shows the “clumps” of L1’s and L2’s that I mentioned in a previous post. In the EM images, they appear as lighter areas, surrounded by darker areas, with each lighter area being the center of a cartridge. In the second image below, the cartridges present something of a “honeycomb” appearance in the EM, in some spots, which makes them really easy to spot…again, each of the honeycomb units being an individual cartridge.
Finally, this link from the FlyWire blog shows all type L cells of a single cartridge together (i.e. L1, L2, L3, L4, and L5). You can add in the T1 cell with Id 720575940619753307 which forms something like a “basket” or “cage” around the cartridge. There are, of course, other cells associated with each cartridge, but those six cells represent the basic structural unit of the cartridges, and should give you a good idea of what you are looking for.
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5667947766874112
Cheers.
Now I clearly see, what you meant by cartridges. I’m working on something else currently, but feel the urge the trace a complete cartridge or two ![]()
This is incredibly helpful, thank you! I look forward to the full guide but I think this is a very useable start to get pecking at the tricky cells that have plagued us.
Incredible work! If y’all need some more L cells to look at here’s a link of a few of them:
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/6701824207749120
1-2 of them are from q/a sheet w/o cells (my cells im looking for their CBs) but other than that they’re all complete. There’s also some retinal axons in there but they synapse and/or ‘Cartridge’ with L cells so I left them in.
a good idea making a guide, in my option it would be really useful to add in pictures that could be used as a printed out version + the 3d links if needed. Since i am tracing on one screen only looking at it on the computer is less useful than having it on paper. And not too many cells at the time since that is making the screen more messy.
Oh, I forgot to mention…
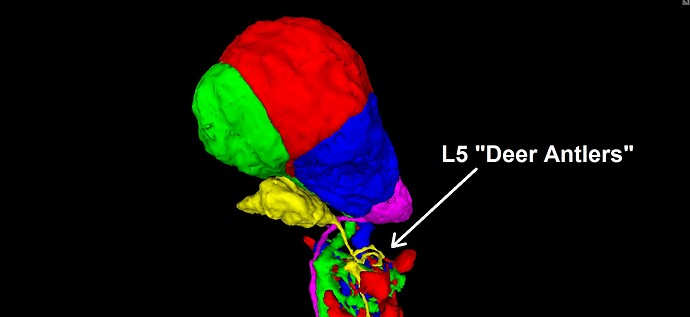
The type L cells form their own well defined and exclusive CB layer, so…I know it goes without saying but, you can always just start randomly clicking on nearby somas, and you will light up L cells all over the place, lol.
The L1, L2, and L3 somas (blue, red, and green below) tend to mush together into a clump, while the L4 and L5 somas (magenta and yellow below) tend to be smaller somas, located just beneath the clump of the first three. Also, the L5 cell seems to always have a small “deer antler” arbor, immediately downstream of the soma, which can help to identify “pinched” L5 segments with the soma attached (have seen a few instances in the Q&A log).
Cheers, all.
https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/4892010485907456
Oh trust u me ik, ive been farming them non stop for 2 weeks by clicking on CBs ![]() hahaha
hahaha
and very often the retinal axons will be within those L ‘cages’. Nicely tightly hugged lol
So, maybe (i aint no scientist lol): Retinal axon > L > T’s > Other cells (info circuit)?
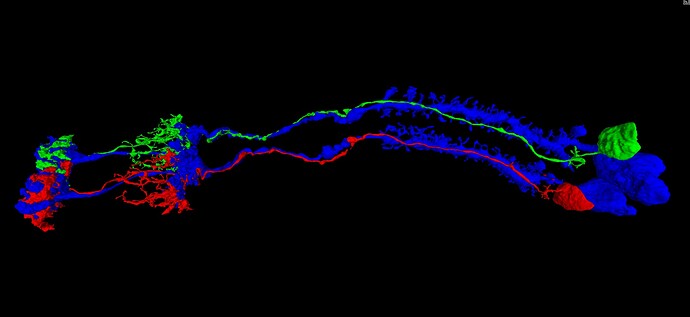
Here’s a ‘full’ circuit (I think), FlyWire and annotated, L’s, T’s, retinal axons, etc. at least for the ‘top’ part, the ‘lower’ part’s a mess of many more types, lol.
Now im wondering if the LAWF’s are taking info from the L’s and T’s and sending it elsewhere and T’s and L’s take info from retinal axons or the pathway is diff.?
the ‘other’ are glia, and other stuff not -attached- to something else larger, standalone branches etc.
Maybe we (the players) should create a sheet in the spreadsheet with all types of neurons listed (glia can be there too ![]() ) and try to find at least one example for each type. It would be both a challenge and a helpful tool for newer players and those less knowledgeable in the field.
) and try to find at least one example for each type. It would be both a challenge and a helpful tool for newer players and those less knowledgeable in the field.
let us do thiiiiiiiiiiiiiiiiiiiiiiiiiissssss!! lol